Abstract
Purpose
To assess the status of EGFR, HER-2, and CCND1 at the gene and protein levels in esophageal squamous cell carcinoma.
Methods
Dual-color FISH assays were performed using DNA probes for EGFR/CEP 7, HER-2/CEP 17, and CCND1/CEP 11. The respective proteins, furthermore, was assessed in IHC assays and correlated with patient and tumor characteristics.
Results
From 55 ESCCs, 8 (15%) tumors showed gene amplification and 20 (36%) had gene overrepresentation (balanced gene and chromosome 7 polysomy) for EGFR. High-level protein expression was frequent (49%), positively correlated with gene copy numbers (kappa=0.4), and associated with well-differentiated histology (p=0.02). For HER-2, gene amplification was detected in a single tumor (2%) and protein overexpression was rare (9%). CCND1 gene was amplified in 23 (42%) tumors; likewise, CCND1 protein overexpression was common (58%) and prevailed in gene overrepresentation or amplification. Only 1 patient showed gene amplification for both EGFR and CCND1. Survival was not associated with EGFR or CCND1 gene/protein status, whereas negative patients for HER-2 protein had a better survival than positive patients (p=0.04).
Conclusions
Frequent overexpression and gene amplification of EGFR and CCND1 make these molecules and their pathways potential therapeutic targets for ESCC. In addition, EGFR and CCND1 appeared to be independently altered suggesting alternative mechanisms for pathway activation. Therapeutic agents targeting these molecules are urged to be tested in clinical trials and comprehensive biological analyses should be included to properly interpret the outcome.
Similar content being viewed by others
Introduction
Esophageal squamous cell carcinoma (ESCC) renders among the most common debilitating malignancies worldwide (Parkin et al. 1997). The outcome of treatment for ESCC has not reached a satisfactory basis due to the incurable disease status at presentation in most patients. The genomic changes involved in the initiation and the progression of esophageal cancer are not very well understood. Classical and molecular cytogenetic techniques have identified multiple genomic regions with losses and gains in copy numbers, clearly supporting the role of major oncogenes and tumor suppressor genes in the tumorigenesis (Schrump and Nguyen 2001)).
The progress in cancer therapy has advanced extensively in the past decade. Amid the cutting edge modalities, molecular-targeted therapy has been a leader. The principle of this modality is to select molecules or pathways essential to the development and/or progression of cancer cells and target them with specifically developed inhibitory agents. Unlike conventional treatment by chemotherapy or radiation, this approach holds a promising clinical efficacy with reduced toxicity to normal cells. Genetic studies using CGH, a powerful molecular cytogenetics technique to survey chromosomal gains and losses in entire genomes, have been performed in over 180 esophageal cancer specimens (Shinomiya et al. 1999; Tada et al. 2000; Stocks et al. 2001; Varis et al. 2001). Among the chromosomal regions commonly identified as showing gain in copy number are 7p12, 17q12, and 11q13, which harbor the genes EGFR, HER-2, and CCND1, respectively. The significance of these three genes in ESCC tumorigenesis has been confirmed using various molecular techniques. By Southern blot analyses, gene amplification of the three targets was detected in a significant fraction of tumors (Adelaide et al. 1995; Suzuki et al. 1997). In addition, proteins encoded by these critical genes have also been examined in ESCC by IHC assays and demonstrated protein expression (Adelaide et al. 1995; Wang 1999); therefore, these genes and the pathways in which they are involved are potential candidates for targeted therapy in ESCC.
Development and progression of ESCC, as of other malignancies, requires a number of key molecules and pathways to function in an orchestrated pattern. Investigations on the status of molecules that play a major role in ESCC tumorigenesis and on how they correlate to each other in the same tumor are necessary to rationalize cancer-targeted therapy. To the best of our knowledge, the simultaneous evaluation of EGFR, HER-2, and CCND1 gene and protein status in ESCC has not been reported before. Additionally, examination of these three genes all together by FISH assay, which is a well-recognized technique to determine the number of copies of the gene in single cells and correlate it with tissue architecture, has not been done. This study was conducted to verify the molecular profiles of these genes in ESCC, individually and in association.
Materials and methods
Specimen selection and processing
Fifty-five consecutive patients with ESCC who underwent esophagectomy from the years 1997 to 2000 at the Prince of Songkla University Hospital in Songkla, Thailand, and whose tumor blocks were determined to be well preserved, were included in this study. Clinicopathological staging and histological grading of tumors followed the current American Joint Committee on Cancer (AJCC) system and the World Health Organization (WHO) classification, respectively. Informed consent was obtained from all patients through standard protocol of the institution and the research project was reviewed and approved by the institutional review board. Selected formalin-fixed, paraffin embedded blocks with >80% of tumor cells were sectioned at 5 μm thickness. Serial sections were stained with H&E, and processed for IHC and dual-color FISH assays addressing EGFR, HER-2, and CCND1.
Dual-color fluorescence in situ hybridization assays
Tissue sections were incubated at 65°C for 4 h, deparaffinized in xylene washes (three times for 10 min), and dehydrated in 100% ethanol. After incubation in 2xSSC (pH 7.0) at 75°C for 23 min, sections were digested with proteinase K (0.25 mg/ml in 2xSSC, pH 7.0) at 45°C for 23 min, rinsed in 2xSSC (pH 7.0) at room temperature for 5 min, and dehydrated in an ethanol series. The following DNA probe sets, all from Vysis Inc. (Downers Grove, Ill.), were used: (a) LSI EGFR (SO)/ CEP 7 (SG); (b) PathVysion LSI HER-2 (SO)/CEP 17 (SG); and (c) LSI CCND1 (SO)/CEP 11 (SG). Each probe set was applied to the selected area on a designated slide. The hybridization areas were covered with glass coverslip and sealed with rubber cement. Co-denaturation of chromosomal and probe DNAs was performed at 80°C for 10 min and hybridization was allowed to occur in a humidified chamber at 37°C for 22 h. Post-hybridization included washes in 1.5 M urea/0.1xSSC (pH 7.0) at 45°C for 30 min and in 2xSSC for 2 min at room temperature. Chromatin counterstaining was achieved with DAPI (0.15 mg/ml in Vectashield Mounting Medium).
Microscopic analyses were performed in epifluorescence microscope equipped with the Quips XL genetic workstation (Applied Imaging, Santa Clara, Calif.). Fluorescence signals were scored using single band filters for DAPI, FITC, and Texas red, a double-band pass filter (FITC and Texas red) and a triple-band pass filter (DAPI, FITC, and Texas Red, Chroma Technology, Brattleboro, Ver.). Histological areas previously selected in the H&E-stained sections were identified in the fluorescence in situ hybridization (FISH)-treated slides. Two hundred non-overlapping interphase nuclei per site were scored for both target gene and control probe signals, following strict scoring guidelines and constant adjustment of microscope focus since signals located at different focal planes. Representative images of each specimen were acquired with a cooled CCD camera (SenSys, Photometrics, Tucson, Ariz.) in monochromatic layers, which were subsequently merged using the SmartCapture software (Vysis, Downers Grove, Ill.).
The following criteria were defined for analysis: For each FISH probe tested, the status of the chromosome used as control was classified as disomy, trisomy, or polysomy when, respectively, on average 2, 3, or more copies were scored per nucleus. FISH patterns were considered balanced when the ratio gene/chromosome copy number per cell was from 0.9–1.2 and unbalanced gene loss was accepted when gene/chromosome ratio was <0.9. Gene amplification was assessed by multiple, clustered copies of the gene in any chromosomal status; it was classified as at low level when gene/chromosome ratio was between 1.3 and 2 and at high level when gene/chromosome ratio was >2. Clusters of signals ranging in number from 3–4 were categorized as small, from 5–9 as medium size, and greater than 9 as large-size clusters. Balanced polysomy was recognized as gene overrepresentation.
Immunohistochemistry assays
To account for the EGFR protein expressions, immunohistochemistry (IHC) assays were performed using the EGFR Mouse Monoclonal Antibody (Zymed, South San Francisco, Calif.) in conditions specified by the manufacturer. Briefly, the tissue sections were deparaffinized in xylene, rehydrated in graded alcohol, and washed in Wash-Buffer solution (Dako). To improve the quality of tissue adhesion, the slides were treated with Poly-L-lysine solution. A positive control using the cell line A431 (human squamous carcinoma, skin), as well as a negative reagent control using the mouse non-specific negative reagent (Dako) was included in each staining run as a reference. The slides were loaded onto an autostainer (Dako) and the following incubations were applied sequentially: 10-min peroxide blocking agent (3%), 10-min Power Block, 10-min proteinase K, 60-min primary EGFR antibody (1:20) or negative control reagent, 30-min ENV+ (Mouse), and 7-min DAB+ substrate-chromogen solution. Buffer rinses were performed following each step except for the DAB+ which required a water rinse. After completion of the auto-staining process, slides were removed from the machine and counterstained with hematoxylin, then rinsed gently in reagent quality water, dehydrated in graded alcohol, cleared in xylene, and coverslipped.
The levels of HER-2 and CCND1 protein expression were evaluated using rabbit anti-human c-erbB-2 oncoprotein (Dako) and mouse anti-human cyclin D1 DCS-6 (Dako) as primary antibodies, respectively. Experiments followed protocols specified by the manufacturer, similar to the description mentioned above. Control specimens were the cell line Calu3 (lung adenocarcinoma) that overexpressed HER-2, and a primary breast carcinoma known to overexpress CCND1. The added-on step was the epitope retrieval process using Target Retrieval Solution (1:10; Dako) at 105°C (Biocare pressure cooker) for 20 min before loading the slides onto the autostainer. ENV+ (rabbit and mouse, for HER-2 and CCND1, respectively) were used as the secondary antibodies and no Power Block was applied.
Analyses of IHC-stained slides were performed using 20× magnification objective in light microscopy. A semi-quantitative criteria was used for EGFR interpretation (Hirsch et al.2003). In detail, the percentage of positive tumor cells per slide (0–100%) was multiplied by the dominant intensity pattern of staining (1=negative or trace; 2=weak; 3=moderate; 4=intense); therefore, the overall score ranged from 0–400. Specimens with overall scores >200, 201–300, and 301–400 were respectively classified as negative/low, with intermediate, and with high levels of expression. For HER-2, Dako’s guidelines for analysis were used. The staining patterns were assessed in four scores: score 0 represents no staining or weak membrane staining in <10% of tumor cells; score 1+ is a faint or barely perceptible partially membrane staining in >10% of tumor cells; score 2+ is a weak to moderate complete membrane staining in >10% of tumor cells; and score 3+ is a strong complete membrane staining in >10% of tumor cells. Only scores 2+ and 3+ were considered to represent protein overexpression. For CCND1, a four-tiered scale following previous publications was applied (Anonymous 2001; Mathew et al. 2001). Score 0 is a negative result with no staining or <10% of cells with nuclear staining; score 1 represents a positive–low tumor with 10–20% stained cells; score 2 is a positive–intermediate tumor with 20–40% stained cells; and score 3 is a positive–high tumor with >40% stained cells. Scores 1–3 represented CCND1 protein overexpression (Mathew et al. 2001).
Statistical analysis
Demographic data available for each patient were age, gender, pathological stage, tumor grade, treatment, and survival outcome. Statistical analyses were carried out with SAS software (SAS Institute, Cary, N.C.). Chi-square, Fisher’s exact, or Mantel-Haenszel linear-by-linear association chi-square tests were used to compare the gene and protein results across levels of patient/tumor characteristics. Kappa statistics were conducted to examine the agreement between gene and protein levels for the three molecular targets. Kappa ≥0.8 indicated excellent agreement, 0.8> kappa ≥0.4 indicated moderate agreement (Stokes et al. 2000). The Kaplan-Meier estimate and log-rank or trend tests were used to examine the survival experience on prognostic factors and targets status.
Results
Patient characteristics
Patient and tumor characteristics are summarized in Table 1. The median age was 63 years (range 43–76 years) and the male:female ratio was 2.7:1. The majority of patients (67%) had stage-3 or stage-4 diseases and most tumors (56%) showed well-differentiated histological grade. All patients underwent esophagectomy and 19 of them also received radiation therapy postoperatively. The median follow-up time was 10.5 months (range 0–61 months). Thirty-nine patients had died, while 13 patients were alive at the time when the study was conducted. Survival data from 3 patients were not available for analysis.
Gene copy number and protein expression
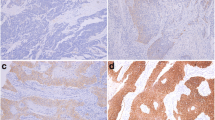
Gene and protein status as well as characteristics for individual patients are shown in Table 2. The number of copies of gene and chromosome per cell were obtained from the FISH assays. According to the criteria described above, the tumors were found to fall into four FISH categories: balanced disomy (BD); balanced trisomy (BT); balanced polysomy (BP); and gene amplification (GA). Unbalanced gene loss was not observed in the 55 tumors and low level of amplification was only detected for HER-2. The four FISH patterns are illustrated in Fig. 1 and the distribution of specimens among these categories for each of the investigated DNA targets are shown in Table 3. Distribution of specimens according to the semiquantitative analysis is presented in Table 4 and negative and overexpressed specimens are illustrated in Fig. 2. The EGFR gene was amplified at high levels in 8 patients (15%) and overrepresented in 20 patients (36%). In tumors with gene amplification, 5 (63%) exhibited medium-sized cluster pattern, whereas the remaining showed large-sized clusters with greater than nine signals (Fig. 1B). EGFR protein was expressed for a high level in 49% of patients. For HER-2, a single patient (2%) showed low level of gene amplification with a medium-sized cluster (Fig. 1D) and 17 patients (31%) had gene overrepresentation; however, the protein was weakly overexpressed (score 2+) in only 5 patients (9%). The CCND1 locus was amplified in 23 tumors (42%), and was overrepresented in 8 tumors (15%). Tumors with CCND1 gene amplification showed heterogeneous cluster patterns: 8 (35%) small sized; 6 (26%) medium sized; and 9 (39%) large-sized clusters (Fig. 1F). Overexpression of CCND1 protein was found in 32 tumors (58%), with 10–20, 20–40, or >40% of expressing cells in 31, 7, and 20% of patients, respectively.
Gene (red signal) and chromosome (green signal) detected by fluorescence in situ hybridization (FISH) in esophageal squamous cell carcinoma (ESCC; probes as described in the text). A Balanced trisomy and B gene amplification for EGFR. C Balanced disomy and D gene amplification for HER-2. E balanced polysomy and F gene amplification for CCND1
The distribution of specimens according to the combination of gene status and protein expression of the three molecular targets is shown in Table 5. The protein expression was dichotomized in low level (IHC negative for HER-2 and CCND1; score ≤300 for EGFR) and high level of expression (IHC positive for HER-2 and CCND1; score >300 EGFR). For EGFR and CCND1, high levels of protein expression prevailed in tumors with gene overrepresentation or amplification (kappa=0.4). Interestingly, a fraction of specimens with gene amplification was found low-level expressed for EGFR (2 tumors) and CCND1 (3 tumors) proteins. For HER-2, no association was detected between gene and protein levels. The rare specimen with weak HER-2 protein overexpression (score 2+) had gene overrepresentation but not clustered amplification and the single specimen with gene amplification has not overexpressed the protein. A similar analysis was performed dichotomizing the results for gene and protein expression, as shown in Fig. 3. Additionally, the gene copy numbers were dichotomized as low (FISH patterns BD+BT) and high (FISH patterns BP+GA) copy numbers per cell. For HER-2, low levels of gene and protein predominated, whereas for EGFR and CCND1, gene overrepresentation and amplification predominated and associated with protein overexpression.
Comparison of dichotomized gene patterns and protein expression levels for EGFR, HER-2, and CCND1 in ESCC. The gene copy numbers were dichotomized as low (FISH patterns BD+BT) and high (FISH patterns BP+GA) copy numbers per cell. The protein expression was dichotomized in low level (IHC negative for HER-2 and CCND1; score ≤300 for EGFR) and high level of expression (IHC positive for HER-2 and CCND1; score >300 for EGFR)
A comparison among the three molecular targets EGFR, HER-2, and CCND1 in each tumor, revealed interesting results. The tumor with HER-2 amplification (patient no. 47) also was amplified for CCND1; however, among the 8 tumors amplified for EGFR and the 23 tumors amplified for CCND1, a single tumor (patient no. 14) was found amplified for both genes. Regarding the protein expression, three tumors (patient nos. 7, 11, and 51) overexpressed all three proteins (Table 2). No association, either at gene or protein levels, was detected between EGFR and HER-2, EGFR and CCND1, or HER-2 and CCND1 in each specimen (kappa <0.4).
Prognostic indicators and survival
EGFR protein overexpression was observed in a higher incidence in tumors with well-differentiated grade (p=0.02). Other than this, no association of EGFR, HER-2, or CCND1 at the gene or the protein level was identified with any of the patient characteristics, including age, gender, survival status, pathological stage, and tumor grade (data given in Table 1). None of these factors, as well as the radiation treatment, was shown to be a significant predictor of survival differences, which precluded their use as prognostic indicators for ESCC. There was no difference in survival experience regarding different levels of EGFR or CCND1 gene or protein expression, individually or combined; however, patients with negative HER-2 protein were found to have a better survival than those with positive protein (p=0.04).
Discussion
The critical role of targeted therapies against molecules involved in the development and progression of neoplasias has been increasingly appreciated. Monoclonal antibodies and tyrosine kinase inhibitors against EGFR and HER-2, and other molecular strategies against their downstream targets including CCND1 are among the leading tested agents (Slichenmyer and Fry 2001); however, some issues yet to be properly addressed before largely applying potential targeted agents into cancer treatment are the lack of method of choice to evaluate target expression, the disparity of results of targets as prognostic factors, and the unclear association between levels of expression and response to targeted therapy (Arteaga 2002). In this study, three molecules were evaluated at protein and gene levels. The number of copies per cell of each gene of interest was evaluated by FISH and correlated with a semi-quantitative protein score determined in IHC assays, using specific antibodies. To the best of our knowledge, a combined evaluation of EGFR, HER-2, and CCND1 gene and protein status has not been reported before for ESCC.
Gain in copy numbers of these genes as well as overexpression of the corresponding proteins appear to play an important role in ESCC. For the EGFR locus, gene overrepresentation, as identified by balanced polysomy of the EGFR gene and chromosome 7, was the prevalent pattern (36%) and gene amplification was found in 15% of patients. These findings agree with previous reports in which EGFR amplification detected by various techniques ranged from 12 to 21% (Kitagawa et al. 1996; Mathew et al. 2001). EGFR protein was commonly expressed for a high level (49%) in this series and this incidence rate falls within the wide range of 19–83% of ESCC overexpressing patients reported previously (Wang et al. 1999; Torzewski et al. 1997). The evaluation of protein and gene levels in the same specimens has shown that high levels of EGFR protein expression prevailed in tumors with gene overrepresentation or amplification supporting the hypothesis that gain in copy number is a critical mechanism for protein overexpression; however, other mechanisms, such as mutations, heterodimerization with HER-2 (cross-talk with HER-2), increased co-expression of ligands, and alterations in molecules that control receptor signaling output (Arteaga 2002) may also account for overexpression. Conversely, changes in HER-2 were less significant in ESCC. Most tumors (67%) had low number of copies of the gene, gene amplification was found in a single tumor, and protein overexpression was rare (9%). These findings are in the lower range of previous publications that described HER-2 amplification in 7–9% and protein overexpression in 0–31% of ESCC (Shiga et al. 1993; Ikeda et al. 1996; Ross and McKenna 2001). Unlike EGFR, HER-2 has shown no association between the gene and protein levels (kappa=−0.1). The small numbers of patients who overexpressed HER-2 protein (Stocks et al. 2001) and who had HER-2 gene amplification (Parkin et al. 1997) most likely explained this result.
In this ESCC series, CCND1 was the gene more frequently amplified (42%) and overrepresented (15%). CCND1 amplification detected by various techniques including FISH and differential polymerase chain reaction assays was in the range of 22–65% (Sheyn et al. 1997; Gramlich et al. 1994). It was also the more heterogeneous patterns for gene amplification with diverse sizes of clusters observed. CCND1 protein was frequently overexpressed (58%), again within the range of frequencies previously described (28–75%) for ESCC patients (Sheyn et al. 1997; Matsumoto et al. 2000). Interestingly, although there was a significant association between gene gain in copy number and protein overexpression, a fraction of tumors with CCND1 gene amplification or overrepresentation did not overexpress the protein indicating that alternative mechanisms controlled protein overexpression.
None of the three targets was found to be associated to each other, either at the gene or protein levels. By far, the CCND1 gene was more frequently amplified and the single tumor with HER-2 amplification was also amplified for CCND1; however, a single tumor shared amplification for both EGFR and CCND1. Similarly, at the protein level, EGFR, HER-2, and CCND1 were concomitantly overexpressed in only three tumors. The amplification and overexpression of EGFR and CCND1 in distinct tumors suggests alternative mechanisms for activation of tumorigenesis pathways.
In our study, we found no evidence favoring gene or protein levels as a better basis for selection of patients who may benefit from targeted therapy. No correlation between EGFR, HER-2, or CCND1 at the gene or protein levels and pathological stage, tumor grade, or radiation treatment was detected. The exception was EGFR, for which the protein level was associated with tumor grade, with higher expression in well-differentiated tumors. No correlation with survival was found for any molecular characteristic, except HER-2 protein. The significance of the HER/erb-B family members as prognostic factors for survival in solid tumors is still under debate, and whether they have a synergystic effect is yet to be addressed (Arteaga 2002). While some studies have shown a correlation between high EGFR or HER-2 expression and poor survival outcome in several cancer types, including esophageal cancer (Arteaga 2002; Nicholson 2001), no impact has also been reported for these tumors (Hirsch et al. 2003; Nicholson et al. 2001; Shimada et al. 1999; Hirsch et al. 2002). CCND1 amplification or expression, on the other hand, was associated with poorer survival outcome in ESCC patients in most series (Stokes et al.20002000; Torzewski et al. 1997). The only significant association with survival in this series was found for patients who overexpressed HER-2 protein and had a poorer survival. The small number of patients in this category (Stocks et al. 2001), however, precludes any clinical conclusion. Constraints in our series for survival analysis could be the small patient population size, the overall poor clinical outcome in these patients, or the insufficiency of effective therapy to render them better responses and improved survival.
Role of these molecules as predictive indicators to therapy has not been extensively addressed. A recent study demonstrated that overexpression of EGFR and CCND1 in ESCC patients treated with chemoradiotherapy correlated with the primary complete response rates in 63 and 50% of patients, respectively (Gotoh et al. 2002). A number of molecular therapeutic agents have been investigated with the ultimate objective of improving survival outcome for patients with various types of solid malignancies. For patients with ESCC whose prognosis is yet to be improved, incorporation of this treatment modality to conventional therapy is a potentially promising strategy. The high levels of EGFR and CCND1 proteins consistently reported in ESCC support the inclusion of the molecular pathways involving these two proteins as therapeutical targets. The higher EGFR expression in well-differentiated tumors might indicate that patients with low-grade tumors will be better responders to EGFR inhibitors.
Conclusion
In conclusion, ESCC are heterogeneous tumors regarding to the profiles of the three molecular markers EGFR, HER-2, and CCND1; however, EGFR and CCND1 pathways seem to be potential candidates for therapeutic targets for ESCC due to their frequent expression. Clinical trials using molecular targeted agents in ESCC are urged and should include comprehensive biological analyses of gene and protein to identify optimal approach for selection of better responders and to properly interpret the results.
References
Adelaide J, Monges G, Derderian C, Seitz JF, Birnbaum D (1995) Oesophageal cancer and amplification of the human cyclin D gene CCND1/PRAD1. Br J Cancer 71:64–68
Anonymous (2001) Prognostic significance of cyclin D1 and E-Cadherin in patients with esophageal squamous cell carcinoma: multiinstitutional retrospective analysis. Research Committee on Malignancy of Esophageal Cancer, Japanese Society for Esophageal Diseases. J Am Coll Surg 192:708–718
Arteaga CL (2002) Epidermal growth factor receptor dependence in human tumors: more than just expression? Oncologist 7 (Suppl 4):31–39
Gotoh M, Kawabe S, Takiuchi K, Katsu I (2002) EGFR and cyclin D1 are possible predictors of sensitivity to chemoradiotherapy (CRT) for esophageal squamous cell carcinoma (SCC). Proc Am Soc Clin Oncol 21:164a
Gramlich TL, Fritsch CR, Maurer D, Eberle M, Gansler TS (1994) Differential polymerase chain reaction assay of cyclin D1 gene amplification in esophageal carcinoma. Diagn Mol Pathol 3:255–259
Hirsch FR, Varella-Garcia M, Franklin WA, Veve R, Chen L, Helfrich B, Zeng C, Baron A, Bunn PA Jr (2002) Evaluation of HER-2/neu gene amplification and protein expression in non-small cell lung carcinomas. Br J Cancer 86:1449–1456
Hirsch FR, Varella-Garcia M, Maria MV di, Veve R, Baron AE, Zeng C, Bunn PAJr, Franklin WA (2003) Epidermal growth factor receptor in non-small cell lung carcinomas: correlation between gene and protein levels and impact on prognosis. J Clin Oncol
Ikeda Y, Ozawa S, Ando N, Kitagawa Y, Ueda M, Kitajima M (1996) Meanings of c-erbB and int-2 amplification in superficial esophageal squamous cell carcinomas. Ann Thorac Surg 62:835–838
Itakura Y, Sasano H, Shiga C, Furukawa Y, Shiga K, Mori S, Nagura H. Epidermal growth factor receptor overexpression in esophageal carcinoma (1994) An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer 74:795–804
Kitagawa Y, Ueda M, Ando N, Ozawa S, Shimizu N, Kitajima M (1996) Further evidence for prognostic significance of epidermal growth factor receptor gene amplification in patients with esophageal squamous cell carcinoma. Clin Cancer Res 2:909–914
Mathew R, Arora S, Khanna R, Shukla NK, Mathur M, Ralhan R (2001) Alterations in cyclin D1 expression in esophageal squamous cell carcinoma in the Indian population. J Cancer Res Clin Oncol 127:251–257
Matsumoto M, Furihata M, Ohtsuki Y, Sasaguri S, Ogoshi S (2000) Immunohistochemical characterization of p57KIP2 expression in human esophageal squamous cell carcinoma. Anticancer Res 20:1947–1952
Nicholson RI, Gee JM, Harper ME (2001) EGFR and cancer prognosis. Eur J Cancer 37 (Suppl 4):S9–S15
Parkin DM, Whelan SL, Ferlay J, Raymond L, Young J (eds) (1997) Cancer incidence in five continents, vol 7. IARC Scientific Publications, Lyon, pp 1066–1071
Ross JS, McKenna BJ (2001) The HER-2/neu oncogene in tumors of the gastrointestinal tract. Cancer Invest 19:554–568
Schrump DS, Nguyen DM (2001) The epidermal growth factor receptor-STAT pathway in esophageal cancer. Cancer J 7:108–111
Sheyn I, Noffsinger AE, Heffelfinger S, Davis B, Miller MA, Fenoglio-Preiser CM (1997) Amplification and expression of the cyclin D1 gene in anal and esophageal squamous cell carcinomas. Hum Pathol 28:270–276
Shiga K, Shiga C, Sasano H, Miyazaki S, Yamamoto T, Yamamoto M, Hayashi N, Nishihira T, Mori S (1993) Expression of c-erbB-2 in human esophageal carcinoma cells: overexpression correlated with gene amplification or with GATA-3 transcription factor expression. Anticancer Res 13:1293–1301
Shimada Y, Imamura M, Watanabe G, Uchida S, Harada H, Makino T, Kano M (1999) Prognostic factors of oesophageal squamous cell carcinoma from the perspective of molecular biology. Br J Cancer 80:1281–1288
Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, Nakamura Y, Inazawa J (1999) Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer 24:337–344
Slichenmyer WJ, Fry DW (2001) Anticancer therapy targeting the erbB family of receptor tyrosine kinases. Semin Oncol 28:67–79
Stocks SC, Pratt N, Sales M, Johnston DA, Thompson AM, Carey FA, Kernohan NM (2001) Chromosomal imbalances in gastric and esophageal adenocarcinoma: specific comparative genomic hybridization-detected abnormalities segregate with junctional adenocarcinomas. Genes Chromosomes Cancer 32:50–58
Stokes ME, Davis CS, Koch GG (2000) Categorical data analysis using the SAS system, 2nd edn. SAS Institute, Cary, North Carolina
Suzuki H, Abo S, Kitamura M, Hashimoto M, Izumi K, Terada K, Sugiyama T (1997) Gene amplification of int-2 and erbB in human esophageal cancer: relationship to clinicopathological variables. Cancer Invest 15:411–415
Tada K, Oka M, Hayashi H, Tangoku A, Oga A, Sasaki K (2000) Cytogenetic analysis of esophageal squamous cell carcinoma cell lines by comparative genomic hybridization: relationship of cytogenetic aberrations to in vitro cell growth. Cancer Genet Cytogenet 117:108–112
Torzewski M, Sarbia M, Verreet P, Bittinger F, Dutkowski P, Heep H, Willers R, Gabbert HE (1997) The prognostic significance of epidermal growth factor receptor expression in squamous cell carcinomas of the oesophagus. Anticancer Res 17:3915–3919
Varis A, Puolakkainen P, Savolainen H, Kokkola A, Salo J, Nieminen O, Nordling S, Knuutila S (2001) DNA copy number profiling in esophageal Barrett adenocarcinoma: comparison with gastric adenocarcinoma and esophageal squamous cell carcinoma. Cancer Genet Cytogenet 127:53–58
Wang LS, Chow KC, Chi KH, Liu CC, Li WY, Chiu JH, Huang MH (1999) Prognosis of esophageal squamous cell carcinoma: analysis of clinicopathological and biological factors. Am J Gastroenterol 94:1933–1940
Acknowledgements. The authors acknowledge the skillful assistance of N. Aumen in the IHC assays, A. Palma in the database entry, and P. Taupradist for reviewing the patient data. This work was supported in part by the NCI grant CCSG P30-CA46934. P. Sunpaweravong and S. Sunpaweravong were post-doctoral fellows sponsored by the Prince of Songkla University, Songkla, Thailand.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sunpaweravong, P., Sunpaweravong, S., Puttawibul, P. et al. Epidermal growth factor receptor and cyclin D1 are independently amplified and overexpressed in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol 131, 111–119 (2005). https://doi.org/10.1007/s00432-004-0610-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-004-0610-7