Abstract
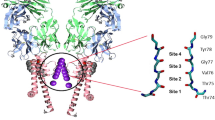
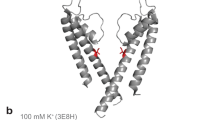
The K+ selectivity filter catalyses the dehydration, transfer and rehydration of a K+ ion in about ten nanoseconds. This physical process is central to the production of electrical signals in biology. Here we show how nearly diffusion-limited rates are achieved, by analysing ion conduction and the corresponding crystallographic ion distribution in the selectivity filter of the KcsA K+ channel. Measurements with K+ and its slightly larger analogue, Rb+, lead us to conclude that the selectivity filter usually contains two K+ ions separated by one water molecule. The two ions move in a concerted fashion between two configurations, K+-water-K+-water (1,3 configuration) and water-K+-water-K+ (2,4 configuration), until a third ion enters, displacing the ion on the opposite side of the queue. For K+, the energy difference between the 1,3 and 2,4 configurations is close to zero, the condition of maximum conduction rate. The energetic balance between these configurations is a clear example of evolutionary optimization of protein function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998).
Eisenman, G., Latorre, R. & Miller, C. Multi-ion conduction and selectivity in the high-conductance Ca++-activated K+ channel from skeletal muscle. Biophys. J. 50, 1025–1034 (1986).
LeMasurier, M., Heginbotham, L. & Miller, C. KcsA: It's a potassium channel. J. Gen. Physiol. 118, 303–314 (2001).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Dunitz, J. D. & Dobler, M. in Biological Aspects of Inorganic Chemistry (eds Addison, A. W., Cullen, W. R., Dolphin, D. & James, B. R.) 113–140 (Wiley, New York, 1977).
Dobler, v. M., Dunitz, J. D. & Kilbourn, B. T. Die struktur des KNCS-Komplexes von nonactin. Helv. Chim. Acta 52, 2573–2583 (1969).
Neupert-Laves, K. & Dobler, M. The crystal structure of a K+ complex of valinomycin. Helv. Chim. Acta 58, 432–442 (1975).
Hodgkin, A. L. & Keynes, R. D. The potassium permeability of a giant nerve fibre. J. Physiol. (Lond.) 128, 61–88 (1955).
Begenisich, T. & De Weer, P. Potassium flux ratio in voltage-clamped squid giant axons. J. Gen. Physiol. 76, 83–98 (1980).
Bestergaard-Bogind, B., Stampe, P. & Christophersen, P. Single-file diffusion through the Ca2+-activated K+ channel of human red cells. J. Membr. Biol. 88, 67–75 (1985).
Neyton, J. & Miller, C. Potassium blocks barium permeation through a calcium-activated potassium channel. J. Gen. Physiol. 92, 549–567 (1988).
Neyton, J. & Miller, C. Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high-conductance Ca2+-activated K+ channel. J. Gen. Physiol. 92, 569–586 (1988).
Alcayaga, C., Cecchi, X., Alvarez, O. & Latorre, R. Streaming potential measurements in Ca2+-activated K+ channels from skeletal and smooth muscle. Biophys. J. 55, 367–371 (1989).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, No. 4. The CCP4 Suite: Programs for X-ray crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Gamblin, S. J., Rodgers, D. W. & Stehle, T. Proc. CCP4 Study Weekend 163–169 (Daresbury Laboratory, Daresbury, 1996).
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Kleywegt, G. J. & Jones, T. A. in From the First Map to Final Model. Proc. CCP4 Study Weekend (eds Bailey, S., Hubbard, R. & Waller, D.) 59–66 (Daresbury Laboratory, Daresbury, 1994).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Kleywegt, G. J. & Jones, T. A. xdlMAPMAN and xdlDATAMAN—Programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D 52, 826–828 (1996).
Heginbotham, L., LeMasurier, M., Kolmakova-Partensky, L. & Miller, C. Single streptomyces lividans K+ channels. Functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 (1999).
Heginbotham, L., Kolmakova-Partensky, L. & Miller, C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998).
Cuello, L. G., Romero, J. G., Cortes, D. M. & Perozo, E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry 37, 3229–3236 (1988).
Koonin, S. E. Computational Physics (Benjamin/Cummings, Menlo Park, 1986).
Kramers, H. A. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica 7, 284–304 (1940).
Kraulis, P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Bacon, D. & Anderson, W. F. A fast algorithm for rendering space filling molecule pictures. J. Mol. Graph 6, 219–220 (1988).
Acknowledgements
We thank, for assistance, the staff at the National Synchrotron Light Source X-25; at Cornell High Energy Synchrotron Source, A1 and F1; at the Advanced Photon Source, ID19; and at the European Synchrotron Radiation Source, ID13. We thank Y. Jiang, R. Dutzler and A. Pico for assistance in data collection; B. Roux for discussions; and F. Valiyaveetil for discussion and advice on the manuscript. This work was supported by a grant from the National Institutes of Health to R.M. R.M. is an investigator in the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Morais-Cabral, J., Zhou, Y. & MacKinnon, R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414, 37–42 (2001). https://doi.org/10.1038/35102000
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35102000
This article is cited by
-
Hierarchically engineered nanochannel systems with pore-in/on-pore structures
NPG Asia Materials (2023)
-
A semiempirical method optimized for modeling proteins
Journal of Molecular Modeling (2023)
-
Switchable Na+ and K+ selectivity in an amino acid functionalized 2D covalent organic framework membrane
Nature Communications (2022)
-
THz trapped ion model and THz spectroscopy detection of potassium channels
Nano Research (2022)
-
Inferring functional units in ion channel pores via relative entropy
European Biophysics Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.