Abstract
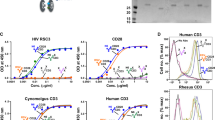
A rapid decline in T-cell counts and the progression to AIDS is often associated with a switch from CCR5-tropic (R5) HIV-1 to CXCR4–tropic (X4) HIV-1 or R5/X4 HIV-1 variants1,2. Experimental infection with R5 HIV-1 causes less T-cell depletion than infection with X4 or R5/X4 variants in T-cell cultures3, in ex vivo infected human lymphoid tissue4,5 and in SCID/hu mice6, despite similar replication levels. Experimental genetic changes in those sequences in gp120 that transform R5 HIV-1 variants into otherwise isogenic X4 viruses make them highly cytopathic6,7. Thus, it is now believed that R5 variants are less cytopathic for T cells than are X4 variants. However, it is not known why CCR5-mediated HIV-1 infection does not lead to a massive CD4+ T-cell depletion, as occurs in CXCR4-mediated HIV-1 infection. Here we demonstrate that R5 HIV-1 isolates are indeed highly cytopathic, but only for CCR5+/CD4+ T cells. Because these cells constitute only a small fraction of CD4+ T cells, their depletion does not substantially change the total CD4+ T-cell count. These results may explain why the clinical stage of HIV disease correlates with viral tropism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tersmette, M. et al. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet 1, 983–985 (1989).
Scarlatti, G. et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nature Med. 3, 1259–1265 (1997).
Berger, E.A. HIV entry and tropism: the chemokine receptor connection. AIDS 11, S3–S16 (1997).
Glushakova, S., Baibakov, B., Margolis, L.B. & Zimmerberg, J. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nature Med. 1, 1320–1322 (1995).
Glushakova, S. et al. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nature Med. 4, 346–349 (1998).
Picchio, G.R., Gulizia, R.J., Wehrly, K., Chesebro, B. & Mosier, D.E. The cell tropism of human immunodeficiency virus type 1 determines the kinetics of plasma viremia in SCID mice reconstituted with human peripheral blood leukocytes. J. Virol. 72, 2002–2009 (1998).
Penn, M.L., Grivel, J.-C, Shramm, B., Godsmith, M.A. & Margolis, L.B. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1- infected human lymphoid tissue. Proc. Natl. Acad. Sci. USA 96, 663–668 (1999).
Bleul, C.C., Wu, L., Hoxie, J.A., Springer, T.A. & Mackay, C.R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94, 1925–1930 (1997).
Zaitseva, M. et al. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nature Med. 3, 1369–1375 (1997).
Glushakova, S., Baibakov, B., Zimmerberg, J. & Margolis, L. Experimental HIV infection of human lymphoid tissue: Correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histoculture inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Retroviruses 13, 461–471 (1997).
Haase, A.T. et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274, 985–989 (1996).
Casella, C.R. & Finkel, T.H. Mechanisms of lymphocyte killing by HIV. Curr. Opin. Hematol. 4, 24–31 (1997).
Bjorndal, A. et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71, 7478–7487 (1997).
Herbein, G. et al. Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature 395, 189–194 (1998).
McDermott, D.H. et al. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352, 866–870 (1998).
Acknowledgements
The authors thank P. Murphy and J. Zimmerberg for many discussions, encouragement and support. This work was supported in part by the NASA/NIH Center for Three-Dimensional Tissue Culture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grivel, JC., Margolis, L. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat Med 5, 344–346 (1999). https://doi.org/10.1038/6565
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/6565
This article is cited by
-
Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection
Nature (2014)
-
HIV integrase and the swan song of the CD4 T cells?
Retrovirology (2013)
-
Use of Human Mucosal Tissue to Study HIV-1 Pathogenesis and Evaluate HIV-1 Prevention Modalities
Current HIV/AIDS Reports (2013)
-
Selective transmission of R5 HIV-1 variants: where is the gatekeeper?
Journal of Translational Medicine (2011)
-
Asymmetric HIV-1 co-receptor use and replication in CD4+ T lymphocytes
Journal of Translational Medicine (2011)