Abstract
Neurofibromatosis type 1 is one of the most common neurocutaneous autosomal dominant disorders. It is caused by mutations in the neurofibromatosis type 1 (NF1) gene and approximately 30–40% of them affect the correct splicing of NF1 pre-mRNA. In this report, we evaluate the effect of five different drugs, previously found to modify splicing in several genetic disorders, on the splicing of mutated NF1 alleles. For this purpose, cell lines derived from patients bearing 19 different NF1-splicing defects were used. Our results showed that kinetin partially corrects the splicing defect in four of the studied mutations (c.910C>T, c.3113G>A, c.6724C>T and c.6791dupA). Our study is a valuable contribution to the field because it identifies new exon-skipping events that can be reversed by kinetin treatment and provides new information about kinetin splicing modulation. However, owing to the nature of mutations in our patients, kinetin treatment could not be used as a therapeutic agent in these cases.
Similar content being viewed by others
Introduction
Neurofibromatosis type 1 (NF1, OMIM#162200) is a common autosomal dominant genetic disorder caused by mutations in the NF1 gene and is mainly characterized by the presence of café-au-lait spots and dermal neurofibromas. More than 1000 different mutations have been uncovered to date,1 and approximately 30–40% of these affect the correct splicing of the gene.1, 2 Recent studies have demonstrated that several drugs can modify transcriptional patterns from alleles carrying splicing mutations in different disorders.3, 4, 5, 6, 7, 8 This modification has mainly been shown in splicing mutations giving a percentage of wild-type transcripts from the mutated allele, which is also described as having a ‘leaky’ effect. In a previous collaborative study in which kinetin corrected aberrant splicing for the most common splicing mutation in familial dysautonomia (FD), we also demonstrated that kinetin was able to restore normal splicing for one NF1 mutation (c.6724C>T, exon 36) in a minigene splicing model,9 which encouraged us to extend our research to other mutations. In this study, we evaluated the effect of kinetin and of four other drugs (valproic acid (VPA), sodium butyrate (SB), (−)-epigallocatechin gallate (EGCG) and aclarubicin), which were previously found to modify splicing in other genetic disorders, in cell lines derived from patients harboring 19 different NF1 splicing mutations to evaluate their putative use as therapeutic agents for NF1.
Materials and methods
A total of 22 patients carrying 19 different NF1 splice mutations were studied (Table 1). Blood samples were collected to obtain both short-term lymphocyte cell cultures and EBV-transformed lymphocyte cell lines from each patient. In two cases, fibroblast cultures from skin or neurofibromas were also obtained. Cell lines were grown as previously described8, 9 and were treated with the following concentrations: kinetin (K, 100 μ M), VPA (50 μ M), SB (5 μg/μl), EGCG (25 μg/ml), all dissolved in water, and aclarubicin (A, 10 nM) dissolved in DMSO (see details in the Results section). Nontreated cell lines or cells treated with DMSO were used as controls. Experiments were performed in duplicate. The relative amount of NF1 full-length and aberrantly spliced transcripts was determined using a semiquantitative RT-PCR approach and an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany)9 (Supplementary Figure 3). Results are shown as percentages of aberrantly spliced transcripts. EBV-transformed lymphocyte cell lines, for which drug treatment elicited an effect on splicing, were treated with 200 μg/ml puromycin (Sigma, St Louis, MO, USA) for 4 h after 20 h of drug treatment to inhibit the nonsense-mediated RNA decay (NMD) mechanism.
Results
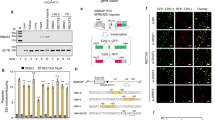
First, experimental conditions were set up in NF1 EBV-transformed cell lines to optimize drug treatment in relation to cellular toxicity and drug concentration, on the basis of previous reports3, 4, 5, 6, 7 (Supplementary Figure 1). Cellular toxicity was only observed for EGCG treatment at 50 μg/ml. Next, short-term lymphocyte cell cultures and EBV-immortalized cell lines from all patients were treated, and no differences were observed between both cell cultures. To compare the results between drugs, data for each patient were normalized by calculating a ratio of aberrantly spliced transcripts from treated cells vs that from untreated cells in each case (Figure 1). A relative decrease, approximately between 30 and 90%, in aberrantly spliced transcripts was observed after kinetin treatment for mutations c.910C>T (exon 7, patients 2 and 3), c.3113G>C (exon 18, patient 11), c.6724C>T (exon 36, patient 16) and c.6791dupA (exon 37, patient 17) in both cell culture types (Figures 1 and 2a and Supplementary Figure 2). It is noteworthy that the observed effect of mutation c.6791dupA (patient 17) is a reduction just above our threshold of 30%; in contrast, no effect was detected for a mutation located close by (c.6792C>A, patients 18–19). Interestingly, cell lines from the patient with mutation c.6724C>T showed less evident exon 36 inclusion that the minigene model from our previous study.9 Differences in the splicing behavior of minigenes and primary cell lines have been reported in literature, which highlights the importance of the surrounding genomic region in determining the splicing.10, 11 Moreover, the heterozygosity of cell lines derived from patients vs homozygosity in constructions should be taken into account when comparing data.
Summary of the different treatments carried out in EBV-transformed lymphocyte cell lines from 22 patients harboring 19 different splicing mutations. Results are given as a ratio of the percentage of aberrantly spliced transcripts in treated cells vs the percentage of aberrant transcripts in untreated cells. X-axis: patients (numbered 1–22, according to Table 1). Y-axis: levels of aberrantly spliced transcripts in treated cells normalized to untreated cells. Slight variations were observed between different patients and different drugs. A clear correction of aberrant splicing was only observed in mutations c.910C>T (patients 2 and 3), c.3113G>C (patient 11), c.6724C>T (patient 16) and c.6791dupA (patient 17) after kinetin treatment. The thick black horizontal line indicates a 30% relative decrease in aberrantly spliced transcripts.
Effect of kinetin treatment on NF1-transcript levels, as well as on aberrantly spliced and full-length transcripts, in EBV-transformed lymphocyte cell lines from patients 3 (c.910C>T), 11 (c.3113G>C), 16 (c.6724C>T) and 17 (c.6791dupA). The effect of different drugs was evaluated before (a) and after (b) puromycin treatment; (c) dose–response after 24 h of treatment using three different concentrations of kinetin; (d) time course at 100 μ M of kinetin treatment. C, untreated cells; K, kinetin (100 μ M); VPA, valproic acid (50 μ M); SB, sodium butyrate (5 μg/μl); EGCG, (−)-epigallocatechin gallate (25 μg/ml); A, aclarubicin (10 nM); DMSO, control of aclarubicin treatment.
To evaluate the effect of kinetin concentration on splicing correction, we performed a dose–response experiment in EBV-immortalized lymphocyte cell lines from all patients showing a kinetin response. Kinetin was tested at 50, 100 and 200 μ M for 24 h, which revealed a concentration-dependent splicing correction, which was incomplete (Figure 2c).
To determine the duration of the kinetin effect on splicing, a time-course experiment was performed at 100 μ M for 1, 4, 8, 16, 24, 48 and 72 h without kinetin replenishment. For mutations c.910C>T, c.3113G>C and c.6791dupA, a partial kinetin effect was observed at 4 h after treatment and was maintained for at least 72 h. For mutation c.6724C>T, the same effect was observed 1 h after treatment and was also maintained for at least 72 h (Figure 2d).
Interestingly, all mutations showing a kinetin effect were nonsense, missense and frameshift mutations that led to exon skipping (Table 1). A previous study by our group demonstrated that mutations in exons affecting correct NF1 splicing produce two types of aberrant transcripts: transcripts with the original mutation and transcripts showing exon skipping.2, 12 Skipping of exons 7, 18 and 37 is in frame, but skipping of exon 36 is out of frame. It is well known that nonsense transcripts could be subjected to degradation by the NMD phenomena. The NMD inhibitor puromycin was added to the treated NF1 cultures to confirm that kinetin acted directly on NF1 mRNA splicing and not on the degradation of nonsense transcripts produced by mutations. Correction of aberrant splicing by kinetin was also observed in cell cultures containing puromycin (Figure 2b).
Finally, we analyzed the effect of kinetin on fibroblast cultures derived from two patients with mutation c.910C>T to evaluate possible differences in distinct cell types. Kinetin treatment was found to have a similar effect on the degree of aberrant splicing correction and the dose-dependent effect (Supplementary Figure 4), indicating that the observed restoration was not restricted to lymphocyte cells.
Discussion
Approximately 30–40% of NF1 patients carry mutations that affect the correct splicing of the NF1 gene. Owing to the high prevalence of these mutations, one of our goals is to develop therapeutic approaches for affected individuals.13 In this regard, several drugs have been found to modify splicing in other genetic conditions.3, 4, 5, 6, 7 In a previous exploratory study, we demonstrated that kinetin improved exon 36 inclusion in a minigene model for NF1 mutation c.6724C>T.9 Here, we evaluated the effect of five different drugs, including kinetin, on 19 different mutations and found that kinetin was able to partially correct aberrant splicing caused by four NF1 splicing mutations (c.910C>T, c.3113G>C, c.6724C>T and c.6791dupA). Exon inclusion was dose dependent, appeared after a few hours and lasted for at least 3 days. The same effect was also observed after inhibition of the NMD mechanism by puromycin treatment, which demonstrates that kinetin acts independently of NMD. The other studied drugs did not show any effect in our set of mutations, which confirms the existence of multiple, distinct mechanisms that contribute to exon selection during pre-mRNA processing.7
Kinetin is a plant cytokinin used in topical applications for its antiaging properties. Its effect on splicing modulation has been demonstrated in IKBKAP, ABI2, BMP2K and NF1 genes.7, 9 Recently, treatment of FD by oral administration of kinetin showed an increase in both IKBKAP-mRNA levels and protein expression.14 Kinetin treatment would be an excellent therapy for FD, in which 99.5% of patients harbor the same splicing mutation, as well as for other splicing disorders in which kinetin could have a modifier effect. Unfortunately, in the case of NF1, the various splicing mutations would hamper the development of a common treatment. Although the mechanism by which kinetin promotes exon inclusion is still unknown, the presence of a CAA motif at the exonic sequence of the 5′ donor site may be involved in splicing regulation.9 However, although this motif is present in exon 36 of the NF1 gene, it is absent in exons 7, 18 and 37, in which a kinetin effect was also observed. This makes it difficult to ascertain the role of the motif in splicing regulation.
In a previous exploratory study, we demonstrated that kinetin improved exon 36 inclusion in a minigene model for NF1 mutation c.6724C>T.9 In cell lines of this patient, we demonstrated that exon 36 inclusion is less evident in cell lines of patients than in our minigene model; this has been previously reported in the literature for other mutations, highlighting the importance of the whole genomic region in the determination of splicing.
In this study, all mutations showing a kinetin effect were exonic. Therefore, correction of aberrant splicing by kinetin treatment produces aberrant transcripts anyway, corresponding to transcripts with the original DNA mutation and, unfortunately, drug treatment will not produce therapeutic effects. However, although our results are not clinically relevant at present, the approach has been demonstrated to be useful for identifying drugs that are able to revert abnormal splicing, and could therefore be of therapeutic interest when skipping-reversion is produced by intronic mutations, as observed in FD,7 or silent mutations, as seen in spinal muscular atrophy.3, 4, 5 Moreover, this type of analysis could also be performed for other emerging drugs or splice factors to identify therapeutic approaches for NF1 patients carrying specific splicing mutations, and would shed some light on NF1 splicing modulation and on the different mechanisms promoting exon inclusion.
References
Messiaen L, Wimmer K : NF1 mutational spectrum; in: Kaufmann D (ed): Neurofibromatoses. Karger: Basel. Monogr Hum Genet 2008; 16: 63–77.
Pros E, Gomez C, Martin T, Fabregas P, Serra E, Lazaro C : Nature and mRNA effect of 282 different NF1 point mutations: focus on splicing alterations. Hum Mutat 2008; 29: E173–E193.
Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H : Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA 2001; 98: 9808–9813.
Andreassi C, Jarecki J, Zhou J et al: Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 2001; 10: 2841–2849.
Brichta L, Hofmann Y, Hahnen E et al: Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet 2003; 12: 2481–2489.
Nissim-Rafinia M, Aviram M, Randell SH et al: Restoration of the cystic fibrosis transmembrane conductance regulator function by splicing modulation. EMBO Rep 2004; 5: 1071–1077.
Slaugenhaupt SA, Mull J, Leyne M et al: Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum Mol Genet 2004; 13: 429–436.
Anderson SL, Qiu J, Rubin BY : EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem Biophys Res Commun 2003; 310: 627–633.
Hims MM, Ibrahim EC, Leyne M et al: Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J Mol Med 2007; 85: 149–161.
Buratti E, Baralle M, Baralle FE : Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res 2006; 34: 3494–3510.
Baralle M, Skoko N, Knezevich A et al: NF1 mRNA biogenesis: effect of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett 2006; 580: 4449–4456.
Pros E, Larriba S, Lopez E et al: NF1 mutation rather than individual genetic variability is the main determinant of the NF1-transcriptional profile of mutations affecting splicing. Hum Mutat 2006; 27: 1104–1114.
Pros E, Fernandez-Rodriguez J, Canet B et al: Antisense therapeutics for neurofibromatosis type 1 caused by deep intronic mutations. Hum Mutat 2009; 30: 454–462.
Gold-von Simson G, Goldberg JD, Rolnitzky LM et al: Kinetin in familial dysautonomia carriers: implications for a new therapeutic strategy targeting mRNA splicing. Pediatr Res 2009; 65: 341–346.
Acknowledgements
We thank all patients, relatives and doctors who participated in this study. C Lázaro wishes to offer special thanks to Drs A Bernards, JF Gusella and S Slaugenhaupt for their continuous support during her sabbatical at MGH (Boston), which inspired this study. This study was supported by grants from the Spanish Health Research Fund: Carlos III Health Institute (BF03/00455, ISCIII-RTICC: RD06/0020/1050 and RD06/0020/1051), Spanish Ministry of Education and Science (SAF2005-00833 and SAF2006-05399) and Catalan Health Institute and Autonomous Government of Catalonia (2009SGR290).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Pros, E., Fernández-Rodríguez, J., Benito, L. et al. Modulation of aberrant NF1 pre-mRNA splicing by kinetin treatment. Eur J Hum Genet 18, 614–617 (2010). https://doi.org/10.1038/ejhg.2009.212
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.212
Keywords
This article is cited by
-
Natural plant hormones cytokinins increase stress resistance and longevity of Caenorhabditis elegans
Biogerontology (2018)
-
Identification of a FUS splicing mutation in a large family with amyotrophic lateral sclerosis
Journal of Human Genetics (2011)