Abstract
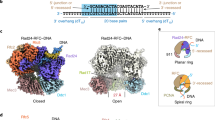
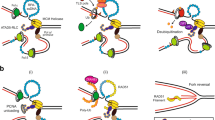
Sliding clamps are ring-shaped proteins that encircle DNA and confer high processivity on DNA polymerases. Here we report the crystal structure of the five-protein clamp loader complex (replication factor-C, RFC) of the yeast Saccharomyces cerevisiae, bound to the sliding clamp (proliferating cell nuclear antigen, PCNA). Tight interfacial coordination of the ATP analogue ATP-γS by RFC results in a spiral arrangement of the ATPase domains of the clamp loader above the PCNA ring. Placement of a model for primed DNA within the central hole of PCNA reveals a striking correspondence between the RFC spiral and the grooves of the DNA double helix. This model, in which the clamp loader complex locks onto primed DNA in a screw-cap-like arrangement, provides a simple explanation for the process by which the engagement of primer–template junctions by the RFC:PCNA complex results in ATP hydrolysis and release of the sliding clamp on DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Huang, C. C., Hearst, J. E. & Alberts, B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J. Biol. Chem. 256, 4087–4094 (1981)
Prelich, G., Kostura, M., Marshak, D. R., Mathews, M. B. & Stillman, B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature 326, 471–475 (1987)
Stukenberg, P. T., Studwell-Vaughan, P. S. & O'Donnell, M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266, 11328–11334 (1991)
Kong, X. P., Onrust, R., O'Donnell, M. & Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, 425–437 (1992)
Krishna, T. S., Kong, X. P., Gary, S., Burgers, P. M. & Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 (1994)
Warbrick, E. The puzzle of PCNA's many partners. Bioessays 22, 997–1006 (2000)
Tsurimoto, T. & Stillman, B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 9, 609–619 (1989)
Tsurimoto, T. & Stillman, B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc. Natl Acad. Sci. USA 87, 1023–1027 (1990)
Lee, S. H., Kwong, A. D., Pan, Z. Q. & Hurwitz, J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase δ. J. Biol. Chem. 266, 594–602 (1991)
Gomes, X. V. & Burgers, P. M. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem. 276, 34768–34775 (2001)
Jeruzalmi, D., O'Donnell, M. & Kuriyan, J. Crystal structure of the processivity clamp loader γ complex of E. coli DNA polymerase III. Cell 106, 429–441 (2001)
Oyama, T., Ishino, Y., Cann, I. K., Ishino, S. & Morikawa, K. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8, 455–463 (2001)
Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. AAA + : A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 (1999)
Guenther, B., Onrust, R., Sali, A., O'Donnell, M. & Kuriyan, J. Crystal structure of the δ′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91, 335–345 (1997)
Yu, R. C., Hanson, P. I., Jahn, R. & Brunger, A. T. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nature Struct. Biol. 5, 803–811 (1998)
Lenzen, C. U., Steinmann, D., Whiteheart, S. W. & Weis, W. I. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell 94, 525–536 (1998)
Goedken, E. R. et al. Fluorescence measurements on the E.coli DNA polymerase clamp loader: implications for conformational changes during ATP and clamp binding. J. Mol. Biol. 336, 1047–1059 (2004)
Ason, B. et al. Mechanism of loading the Escherichia coli DNA polymerase III β sliding clamp on DNA. Bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J. Biol. Chem. 278, 10033–10040 (2003)
Heras, B. et al. Dehydration converts DsbG crystal diffraction from low to high resolution. Structure (Camb.) 11, 139–145 (2003)
Shamoo, Y. & Steitz, T. A. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99, 155–166 (1999)
Gulbis, J. M., Kelman, Z., Hurwitz, J., O'Donnell, M. & Kuriyan, J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 (1996)
Lee, S. et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 (2003)
Niwa, H., Tsuchiya, D., Makyio, H., Yoshida, M. & Morikawa, K. Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure (Camb.) 10, 1415–1423 (2002)
Menz, R. I., Walker, J. E. & Leslie, A. G. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106, 331–341 (2001)
Yang, W., Gao, Y. Q., Cui, Q., Ma, J. & Karplus, M. The missing link between thermodynamics and structure in F1-ATPase. Proc. Natl Acad. Sci. USA 100, 874–879 (2003)
Waga, S. & Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 (1998)
Saenger, W. Principles of Nucleic Acid Structure (Springer, New York, 1984)
Xiong, Y. & Sundaralingam, M. Crystal structure of a DNA RNA hybrid duplex with a polypurine RNA r(gaagaagag) and a complementary polypyrimidine DNA d(CTCTTCTTC). Nucleic Acids Res. 28, 2171–2176 (2000)
Jones, S., van Heyningen, P., Berman, H. M. & Thornton, J. M. Protein–DNA interactions: A structural analysis. J. Mol. Biol. 287, 877–896 (1999)
Jeruzalmi, D. et al. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106, 417–428 (2001)
Story, R. M., Weber, I. T. & Steitz, T. A. The structure of the E. coli recA protein monomer and polymer. Nature 355, 318–325 (1992)
Hortnagel, K. et al. Saturation mutagenesis of the E. coli RecA loop L2 homologous DNA pairing region reveals residues essential for recombination and recombinational repair. J. Mol. Biol. 286, 1097–1106 (1999)
Mirshad, J. K. & Kowalczykowski, S. C. Biochemical characterization of a mutant RecA protein altered in DNA-binding loop 1. Biochemistry 42, 5945–5954 (2003)
Notarnicola, S. M., Park, K., Griffith, J. D. & Richardson, C. C. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 270, 20215–20224 (1995)
Washington, M. T., Rosenberg, A. H., Griffin, K., Studier, F. W. & Patel, S. S. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 271, 26825–26834 (1996)
Skordalakes, E. & Berger, J. M. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003)
Sawaya, M. R., Guo, S., Tabor, S., Richardson, C. C. & Ellenberger, T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99, 167–177 (1999)
Singleton, M. R., Sawaya, M. R., Ellenberger, T. & Wigley, D. B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000)
Tsurimoto, T. & Stillman, B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer–template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 266, 1950–1960 (1991)
Huang, H., Chopra, R., Verdine, G. L. & Harrison, S. C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669–1675 (1998)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Gonzalez, A. Optimizing data collection for structure determination. Acta Crystallogr. D 59, 1935–1942 (2003)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
La Fortelle, E. D. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 276, 476–494 (1997)
Collaborative Computational Project Number 4, The CCP4 suite programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Kleywegt, G. J. & Jones, T. A. Efficient rebuilding of protein structures. Acta Crystallogr. D 50, 829–832 (1996)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Lu, G. TOP: A new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr. 33, 176–183 (2000)
Miyata, T. et al. Direct view of the clamp-loading complex for processive DNA replication. Nature Struct. Mol. Biol. (in the press)
Acknowledgements
We thank J. Berger, E. Goedken, M. Hingorani, D. Jeruzalmi, A. Johnson, S. Kazmirski, I. Nodelman, M. Podobnik, N. Yao and M. Young for scientific discussions, S. Chung and J. Finkelstein for technical assistance, K. Morikawa for sharing data prior to publication, L. Leighton for help with figure preparation, D. King for mass spectroscopy analysis, and members of the Kuriyan laboratory for support and helpful discussions. We appreciate the assistance of C. Ralston, G. McDermott and the scientific staff of the Advanced Light Source (LBNL, Berkeley) in data collection. This work was supported by grants from the National Institutes of Health (J.K. and M.O.D.) and a Ruth L. Kirschstein National Research Service Award through the National Institute of General Medical Sciences (G.D.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Removal of the RFC “arginine finger” residues severely compromises the RFC ATPase. (JPG 18 kb)
Supplementary Figure 2
Experimental MAD-phased electron density maps surrounding bound nucleotides reveal the triphosphate character for RFC-A, B, C and D. (JPG 70 kb)
Supplementary Figure 3
The A:B interface of the RFC complex is more closely packed than those of NSF and HslU. (JPG 77 kb)
Supplementary References
References for supplementary methods and figures. (DOC 20 kb)
Rights and permissions
About this article
Cite this article
Bowman, G., O'Donnell, M. & Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature 429, 724–730 (2004). https://doi.org/10.1038/nature02585
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02585
This article is cited by
-
Deciphering functional roles of protein succinylation and glutarylation using genetic code expansion
Nature Chemistry (2024)
-
Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM
Nature Structural & Molecular Biology (2024)
-
Synergism between CMG helicase and leading strand DNA polymerase at replication fork
Nature Communications (2023)
-
Identification of DNA damage response-related genes as biomarkers for castration-resistant prostate cancer
Scientific Reports (2023)
-
Unchanged PCNA and DNMT1 dynamics during replication in DNA ligase I-deficient cells but abnormal chromatin levels of non-replicative histone H1
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.