Abstract
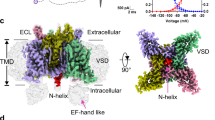
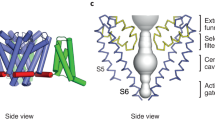
In excitable cells, voltage-gated sodium (NaV) channels activate to initiate action potentials and then undergo fast and slow inactivation processes that terminate their ionic conductance1,2. Inactivation is a hallmark of NaV channel function and is critical for control of membrane excitability3, but the structural basis for this process has remained elusive. Here we report crystallographic snapshots of the wild-type NaVAb channel from Arcobacter butzleri captured in two potentially inactivated states at 3.2 Å resolution. Compared to previous structures of NaVAb channels with cysteine mutations in the pore-lining S6 helices (ref. 4), the S6 helices and the intracellular activation gate have undergone significant rearrangements: one pair of S6 helices has collapsed towards the central pore axis and the other S6 pair has moved outward to produce a striking dimer-of-dimers configuration. An increase in global structural asymmetry is observed throughout our wild-type NaVAb models, reshaping the ion selectivity filter at the extracellular end of the pore, the central cavity and its residues that are analogous to the mammalian drug receptor site, and the lateral pore fenestrations. The voltage-sensing domains have also shifted around the perimeter of the pore module in wild-type NaVAb, compared to the mutant channel, and local structural changes identify a conserved interaction network that connects distant molecular determinants involved in NaV channel gating and inactivation. These potential inactivated-state structures provide new insights into NaV channel gating and novel avenues to drug development and therapy for a range of debilitating NaV channelopathies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hodgkin, A. L. & Huxley, A. F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. (Lond.) 117, 500–544 (1952)
Rudy, B. Slow inactivation of the sodium conductance in squid giant axons. Pronase resistance. J. Physiol. (Lond.) 283, 1–21 (1978)
Vilin, Y. Y. & Ruben, P. C. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem. Biophys. 35, 171–190 (2001)
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011)
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007)
Ren, D. et al. A prokaryotic voltage-gated sodium channel. Science 294, 2372–2375 (2001)
Catterall, W. A. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26, 13–25 (2000)
Armstrong, C. M., Bezanilla, F. & Rojas, E. Destruction of sodium conductance inactivation in squid axons perfused with pronase. J. Gen. Physiol. 62, 375–391 (1973)
West, J. W. et al. A cluster of hydrophobic amino acid residues required for fast Na+ channel inactivation. Proc. Natl Acad. Sci. USA 89, 10910–10914 (1992)
Eaholtz, G., Scheuer, T. & Catterall, W. A. Restoration of inactivation and block of open sodium channels by an inactivation gate peptide. Neuron 12, 1041–1048 (1994)
Nesterenko, V. V., Zygmunt, A. C., Rajamani, S., Belardinelli, L. & Antzelevitch, C. Mechanisms of atrial-selective block of Na+ channels by ranolazine: II. Insights from a mathematical model. Am. J. Physiol. Heart Circ. Physiol. 301, H1615–H1624 (2011)
Wang, Y. et al. Merging structural motifs of functionalized amino acids and α-aminoamides results in novel anticonvulsant compounds with significant effects on slow and fast inactivation of voltage-gated sodium channels and in the treatment of neuropathic pain. ACS Chem. Neurosci. 2, 317–322 (2011)
Pavlov, E. et al. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys. J. 89, 232–242 (2005)
Yue, L., Navarro, B., Ren, D., Ramos, A. & Clapham, D. E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 120, 845–853 (2002)
Zhao, Y., Yarov-Yarovoy, V., Scheuer, T. & Catterall, W. A. A gating hinge in Na+ channels: a molecular switch for electrical signaling. Neuron 41, 859–865 (2004)
Zhao, Y., Scheuer, T. & Catterall, W. A. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc. Natl Acad. Sci. USA 101, 17873–17878 (2004)
Irie, K. et al. Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J. Biol. Chem. 285, 3685–3694 (2010)
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998)
Cuello, L. G., Jogini, V., Cortes, D. M. & Perozo, E. Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208 (2010)
Chen, Y., Yu, F. H., Surmeier, D. J., Scheuer, T. & Catterall, W. A. Neuromodulation of Na+ channel slow inactivation via cAMP-dependent protein kinase and protein kinase C. Neuron 49, 409–420 (2006)
Wang, S. Y. & Wang, G. K. A mutation in segment I-S6 alters slow inactivation of sodium channels. Biophys. J. 72, 1633–1640 (1997)
Vilin, Y. Y., Fujimoto, E. & Ruben, P. C. A single residue differentiates between human cardiac and skeletal muscle Na+ channel slow inactivation. Biophys. J. 80, 2221–2230 (2001)
Zarrabi, T. et al. A molecular switch between the outer and the inner vestibules of the voltage-gated Na+ channel. J. Biol. Chem. 285, 39458–39470 (2010)
Ragsdale, D. S., McPhee, J. C., Scheuer, T. & Catterall, W. A. Molecular determinants of state-dependent block of sodium channels by local anesthetics. Science 265, 1724–1728 (1994)
Yarov-Yarovoy, V. et al. Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na+ channel α subunit. J. Biol. Chem. 276, 20–27 (2001)
Yarov-Yarovoy, V. et al. Role of amino acid residues in transmembrane segments IS6 and IIS6 of the sodium channel α subunit in voltage-dependent gating and drug block. J. Biol. Chem. 277, 35393–35401 (2002)
Nau, C. & Wang, G. K. Interactions of local anesthetics with voltage-gated Na+ channels. J. Membr. Biol. 201, 1–8 (2004)
Hille, B. Local anesthetics: hydrophilic and hydrophobic pathways for the drug-receptor reaction. J. Gen. Physiol. 69, 497–515 (1977)
Villalba-Galea, C. A., Sandtner, W., Starace, D. M. & Bezanilla, F. S4-based voltage sensors have three major conformations. Proc. Natl Acad. Sci. USA 105, 17600–17607 (2008)
Wang, J. et al. Mapping the receptor site for α-scorpion toxins on a Na+ channel voltage sensor. Proc. Natl Acad. Sci. USA 108, 15426–15431 (2011)
Koth, C. M. & Payandeh, J. Strategies for the cloning and expression of membrane proteins. Adv. Protein Chem. Struct. Biol. 76, 43–86 (2009)
Faham, S. et al. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 14, 836–840 (2005)
Kauffmann, B., Weiss, M. S., Lamzin, V. S. & Schmidt, A. How to avoid premature decay of your macromolecular crystal: a quick soak for long life. Structure 14, 1099–1105 (2006)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006)
Winn, M. D., Murshudov, G. N. & Papiz, M. Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Laskowski, R. A., Moss, D. S. & Thornton, J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 (1993)
Petrek, M., Kosinova, P., Koca, J. & Otyepka, M. MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15, 1357–1363 (2007)
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008)
Kleywegt, G. J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D 52, 842–857 (1996)
PyMOL Molecular Graphics System . Version 1.2r3pre (Schrödinger LLC).
Acknowledgements
We thank B. Hille for comments on a draft of the manuscript and members of the Zheng and Catterall groups for their insight and support throughout this project. We are grateful to the beamline staff at the Advanced Light Source (BL8.2.1 and BL8.2.2) for their assistance during data collection. J.P. acknowledges support from a Canadian Institutes of Health Research fellowship and the support of N. and E. Payandeh. This work was supported by grants from the National Institutes of Health (R01 NS15751 and U01 NS058039 to W.A.C.) and by the Howard Hughes Medical Institute (N.Z.). We dedicate this work to the memory of our colleague Laura Sheard.
Author information
Authors and Affiliations
Contributions
W.A.C. and N.Z. are both senior authors. J.P., N.Z. and W.A.C. conceived and J.P. conducted the protein purification and crystallization experiments. J.P. and N.Z. determined and analysed the structure of NaVAb. T.M.G., T.S. and W.A.C. conceived, T.M.G. conducted, and T.M.G., T.S. and W.A.C. analysed the electrophysiology experiments. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary References, Supplementary Tables 1-3 and Supplementary Figures 1-10. (PDF 8111 kb)
Rights and permissions
About this article
Cite this article
Payandeh, J., Gamal El-Din, T., Scheuer, T. et al. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139 (2012). https://doi.org/10.1038/nature11077
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11077
This article is cited by
-
The structural basis of divalent cation block in a tetrameric prokaryotic sodium channel
Nature Communications (2023)
-
Cannabidiol inhibits Nav channels through two distinct binding sites
Nature Communications (2023)
-
Voltage sensors of a Na+ channel dissociate from the pore domain and form inter-channel dimers in the resting state
Nature Communications (2023)
-
Quaternary structure independent folding of voltage-gated ion channel pore domain subunits
Nature Structural & Molecular Biology (2022)
-
Focused ion beam milling based formation of nanochannels in silicon-glass microfluidic chips for the study of ion transport
Microfluidics and Nanofluidics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.