Abstract
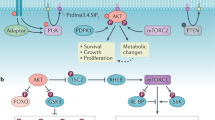
The YAP and TAZ mediators of the Hippo pathway (hereafter called YAP/TAZ) promote tissue proliferation and organ growth. However, how their biological properties intersect with cellular metabolism remains unexplained. Here, we show that YAP/TAZ activity is controlled by the SREBP/mevalonate pathway. Inhibition of the rate-limiting enzyme of this pathway (HMG-CoA reductase) by statins opposes YAP/TAZ nuclear localization and transcriptional responses. Mechanistically, the geranylgeranyl pyrophosphate produced by the mevalonate cascade is required for activation of Rho GTPases that, in turn, activate YAP/TAZ by inhibiting their phosphorylation and promoting their nuclear accumulation. The mevalonate–YAP/TAZ axis is required for proliferation and self-renewal of breast cancer cells. In Drosophila melanogaster, inhibition of mevalonate biosynthesis and geranylgeranylation blunts the eye overgrowth induced by Yorkie, the YAP/TAZ orthologue. In tumour cells, YAP/TAZ activation is promoted by increased levels of mevalonic acid produced by SREBP transcriptional activity, which is induced by its oncogenic cofactor mutant p53. These findings reveal an additional layer of YAP/TAZ regulation by metabolic cues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Stanger, B. Z. Organ size determination and the limits of regulation. Cell Cycle 7, 318–324 (2008).
Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Cordenonsi, M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011).
Chen, D. et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature Med. 18, 1511–1517 (2012).
Sudol, M. Newcomers to the WW domain-mediated network of the Hippo tumor suppressor pathway. Genes Cancer 1, 1115–1118 (2010).
Piccolo, S., Cordenonsi, M. & Dupont, S. Molecular pathways: YAP and TAZ take center stage in organ growth and tumorigenesis. Clin. Cancer Res. 19, 4925–4930 (2013).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012).
Demierre, M. F., Higgins, P. D., Gruber, S. B., Hawk, E. & Lippman, S. M. Statins and cancer prevention. Nature Rev. Cancer 5, 930–942 (2005).
Larsson, O. HMG-CoA reductase inhibitors: role in normal and malignant cells. Crit. Rev. Oncol. Hematol. 22, 197–212 (1996).
Clendening, J. W. & Penn, L. Z. Targeting tumor cell metabolism with statins. Oncogene 31, 4967–4978 (2012).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Goldstein, J. L. & Brown, M. S. Regulation of the mevalonate pathway. Nature 343, 425–430 (1990).
Wada, K., Itoga, K., Okano, T., Yonemura, S. & Sasaki, H. Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914 (2011).
Sansores-Garcia, L. et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325–2335 (2011).
Casey, P. J. & Seabra, M. C. Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292 (1996).
Keller, P. J., Fiordalisi, J. J., Berzat, A. C. & Cox, A. D. Visual monitoring of post-translational lipid modifications using EGFP-GTPase probes in live cells. Methods 37, 131–137 (2005).
Zhao, B., Li, L., Lei, Q. & Guan, K. L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 (2010).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Freed-Pastor, W. A. et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 (2012).
Nielsen, S. F., Nordestgaard, B. G. & Bojesen, S. E. Statin use and reduced cancer-related mortality. N. Engl. J. Med. 368, 576–577 (2013).
Lei, Q. Y. et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell Biol. 28, 2426–2436 (2008).
Ren, F., Zhang, L. & Jiang, J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol. 337, 303–312 (2010).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Brown, M. S. & Goldstein, J. L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997).
Parsons, R. B. et al. Statins inhibit the dimerization of β-secretase via both isoprenoid- and cholesterol-mediated mechanisms. Biochem. J. 399, 205–214 (2006).
Kamisuki, S. et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 ( 2009).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Zhang, H. et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284, 13355–13362 (2009).
Girardini, J. E. et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20, 79–91 (2011).
Wang, Z. et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc. Natl Acad. Sci. USA 111, E89-9 (2014).
Clendening, J. W. et al. Dysregulation of the mevalonate pathway promotes transformation. Proc. Natl Acad. Sci. USA 107, 15051–15056 (2010).
Rustighi, A. et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol. Med. 6, 99–119 (2014).
Acknowledgements
We thank A. Comel, F. Mantovani, A. Rustighi and D. Walerich for helpful discussion and reading the manuscript and A. Testa for discussions and proofreading the manuscript. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and AIRC Special Program Molecular Clinical Oncology ‘5 per mille’ to S.P., A.R. and G.D.S. and the Italian Ministry of Education, University and Research (COFIN, FIRB-accordi di programma 2010 cod.RBAP10XKNC_003) to G.D.S. M.M. is supported by the FIRB RBAP11Z4Z9 project from the Italian Ministry of Education, University and Research. V.S. is supported by grant FIRB no. RBFR10V8K6 from the Italian Ministry of Education, University and Research. G.S. is a fellow of the Fondazione Italiana per la Ricerca sul Cancro (FIRC). We acknowledge G. Pastore for technical support and F. Vita for technical support with electron microscopy. The results published here are in part based on data generated by The Cancer TCGA Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov.
Author information
Authors and Affiliations
Contributions
G.S., N.R., E.I., A.M., M.C. and S.D. performed the experiments. V.S. performed the Drosophila experiments. R.S. performed mice experiments. M.M. performed the high-content screening. A.R. supervised R.S. S.P. performed the bioinformatic analysis. G.S., S.P. and G.D.S. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Statins inhibit YAP/TAZ nuclear localization.
(a) Schematic representation of the high-content screening. MDA-MB-231 cells were seeded in 384-well plates and 24 h later the FDA-approved compounds were added to cells at 1 or 10 μM. 24 h after treatment, cells were fixed and processed for immunofluorescence for YAP/TAZ and stained with Hoechst. Automated image acquisition and analysis was then performed to analyse the subcellular localization of YAP/TAZ. The screening was performed in duplicate; ca. 4,500 cells were analysed per experimental condition and replicate. (b) Correlation between the two screening replicates at 10 μM. Dashed lines represent the levels of cells treated with DMSO. n = 2 biological replicates. (c) Representative images from the screening. MDA-MB-231 stained for Hoechst and YAP/TAZ after treatment with DMSO or the five statins present in the library are shown. Representative images are shown. Experiment repeated two times. Scale bars, 100 μm. (d) Cells were treated with DMSO (NT) or Simvastatin (SIM) 1 μM alone or with mevalonic acid 0.5 mM for 24 h before fixation. Left: representative images of immunofluorescence in MDA-MB-231. Scale bars, 15 μm. Right: quantification of cells with nuclear YAP/TAZ. Representative images are shown. Experiment repeated four times. Data are derived from n = 4 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (e) Western blot showing nuclear-cytoplasmic fractionation of MDA-MB-231 cells after treatment with Cerivastatin 1 μM alone or with mevalonic acid 0.5 mM for 24 h. Experiment repeated three times. Scale bars, 15 μm. *P < 0.05, **P < 0.01; Students t-test is used throughout.
Supplementary Figure 2 Bisphosphonates and geranylgeranyl transferase inhibitors block YAP/TAZ nuclear localization.
(a) qPCR and luciferase reporter assay (8XGTII-lux) to measure YAP/TAZ transcriptional activity. MDA-MB-231 cells were transfected with control siRNA (siCTL) or siRNA against HMG-CoA Reductase (siHMGCR). 72 h after transfection cells were collected and analysed for mRNA expression (left) or luciferase activity (right). Data were normalized to siCTL. Error bars represent mean ± s.d., from n = 3 biological replicates. (b) Representative images of YAP/TAZ in MDA-MB-231 cells after treatment with inhibitors: Zoledronic Acid (ZA) (50 μM), FTI-277 (1 μM), YM-53601 (1 μM), GGTI-298 (1 μM) for 24 h. Experiment repeated four times. Data are derived from three independent experiments where at least 300 cells were scored. Scale bars, 15 μm. (c) Luciferase reporter assay (8XGTII-lux) to measure YAP/TAZ transcriptional activity. MDA-MB-231 cells were treated with DMSO (NT) or Geranylgeranyl transferase inhibitor (GGTI-298) 1 μM for 24 h. Data were normalized to NT. Error bars represent mean ± s.d., from n = 4 biological replicates. (d) Immunofluorescence images shown in Fig. 2d, here presented with their nuclear staining (Hoechst) and zoom. Scale bars, 15 μm. (e) Cells were treated with DMSO (NT) or Cerivastatin (CER) 1 μM alone or with farnesyl phyrophosphate (FPP) or Squalene (SQ) for 24 h before fixation. Experiment repeated four times. Data are derived from n = 4 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d.;. (f) MDA-MB-231 cells were transfected with luciferase 8XGTII-lux reporter. After 24 h cells were treated with DMSO (NT) or Zoledronic Acid (ZA) alone or with GGPP for 24 h. Data were normalized to NT. Error bars represent mean ± s.d., from n = 4 biological replicates. (g) Western blot of MDA-MB-231 cells after treatment with DMSO (NT) or Zoledronic Acid (ZA) for 24 h. Representative blots are shown. Experiment repeated three times. (h) qPCR analysis in MDA-MB-231 transfected with indicated siRNAs. Error bars represent mean ± s.d., from n = 3 biological replicates. (i) Parental (CTL), siRNA-resistant WT-YAP and 5SA-YAP-overexpressing MDA-MB-231 cells were transfected with a combination of YAP/TAZ siRNA (siYAP/TAZ). The day after, cells were treated with Cerivastatin for 24 h and analysed by immunofluorescence. Representative images are shown. Experiment repeated four times. Data are derived from three independent experiments where at least 300 cells were scored. Scale bars, 15 μm. *P < 0.05, **P < 0.01; Students t-test is used throughout.
Supplementary Figure 3 Statins inhibit Rho-GTPases localization and activity by reducing the levels of GGPP.
(a) Quantification of H1299 cells with nuclear GFP-RhoA after treatment with DMSO (NT) or Cerivastatin (CER) 1 μM alone or with geranylgeranyl pyrophosphate (GGPP) 20 μM for 24 h. Data are derived from n = 3 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (b) Cytoplasmic fraction of H1299 cells after treatment with DMSO (NT) or Cerivastatin (CER) 1 μM alone or with geranylgeranyl pyrophosphate (GGPP) 20 μM for 24 h was analysed by western blot. Representative blots are shown. Experiment repeated three times. (c) G-actin/F-actin assay on MCF10A cells. Cells were treated over day with the indicated drugs, lysed and centrifuged in F-actin stabilization buffer to quantify the relative amount of filamentous actin (F-actin) content in the pellet (P) versus free globular actin (G-actin) content in the supernatant (S). LatrunculinA served as positive control for F-actin dissociation. Phalloidin for F-actin stabilization. Representative blots are shown. Experiment repeated three times. (d) Immunofluorescence images shown in Fig. 3a, here presented with their nuclear staining (Hoechst). Scale bars, 15 μm. (e) Quantification of GTP-bound RhoA together with total cell lysates was assessed by western blot after Rhotekin pull-down assay. Cells were treated with DMSO (NT) or with Cerivatsatin (CER) 1 μM alone or with geranylgeranyl pyrophosphate 20 μM (GGPP+CER) or mevalonic acid (MVA+CER) 0.5 mM for 24 h. Ratio was calculated by densitometric analysis of western blot. Representative blots are shown. Experiment repeated three times. (f) Immunofluorescence images shown in Fig. 3c, here presented with their nuclear staining (Hoechst). Subcellular localization images of transiently transfected GFP-RhoA and GFP-RhoA-F in H1299 cells after treatment with FTI-277 1 μM for 24 h were added. Scale bars, 15 μm. *P < 0.05, **P < 0.01; Students t-test is used throughout.
Supplementary Figure 4 Statins and GGTI inhibit YAP/TAZ through Rho-GTPases and show tumor-suppressor activities.
(a) Quantification of cells with nuclear GFP-RhoA-F in MDA-MB-231 cells stably expressing the GFP-RhoA-F construct after treatment with DMSO (NT) or with Cerivastatin (CER) 1 μM. Data are derived from n = 3 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (b) Quantification of cells with nuclear YAP/TAZ in MDA-MB-231 cells stably expressing the GFP-RhoA-F construct after treatment with DMSO (NT) or with Cerivastatin (CER) 1 μM. Data are derived from n = 4 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (c) qPCR analysis in MDA-MB-231 stably expressing the GFP-RhoA-F construct. Cells were treated with DMSO (NT) or Cerivastatin (CER) 1 μM alone or with mevalonic acid 0.5 mM for 48 h. Error bars represent mean ± s.d., from n = 3 biological replicates. (d) Percentage of H1299 cells with nuclear YAP/TAZ. Cells stably expressing the indicated constructs were treated with DMSO (NT) or GGTI-298 (1 μM) for 24 h. Data are derived from n = 3 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (e) Luciferase reporter assay (8XGTII-lux). Cells were treated as in d. Data are normalized to NT. Error bars represent mean ± s.d., from n = 3 biological replicates. (f) Viability assay (ATPlite) of control and YAP-5SA stably expressing MDA-MB-231 cells after treatment with increasing amount of Cerivastatin (0, 0.1, 1, 10 μM and 10 μM with mevalonic acid 0.5 mM) for 48 h. Data are normalized to NT. Error bars represent mean ± s.d., from n = 3 biological replicates. (g) Viability assay (WST-1) of control and YAP-5SA stably expressing MCF10A MII cells after treatment with increasing amount of Cerivastatin (0, 0.1, 1, 10 μM and 10M with mevalonic acid 0.5 mM) for 48 h. Data are normalized to NT. Error bars represent mean ± s.d., from n = 3 biological replicates. (h) Control and YAP-5SA stably expressing MCF10A MII cells were treated with increasing amount of Cerivastatin (0, 0.1, 1, 10 μM) for 48 h. Cleaved PARP was detected by western blot. Representative blots are shown. Experiment repeated three times. (i) Clonogenic assay on MDA-MB-231 cells. Cells were transfected with the indicated siRNA. After 1 day cells were seeded in 10-cm petri dishes. Growing colonies were fixed after one week and stained with crystal violet. Experiment repeated two times. Scale bar, 2 cm. (j) To evaluate the intra-tumoral effect of zoledronic acid on the mevalonate pathway, total RNA was extracted from tumors from control (saline) or zoledronic acid (ZA) treated mice and the mRNA levels of the mevalonate pathway target gene LDLR were determined by qPCR. n = 5 mice. *P < 0.05, **P < 0.01; Students t-test is used throughout.
Supplementary Figure 5 GGTI inhibits cell migration by reducing YAP activity.
(a) MCF10A MII cells expressing vector (CTL), YAP wild type (WT), and YAP-5SA (5SA) were analysed for migration by a wound-healing assay after treatment with DMSO (NT) or GGTI-298 1 μM for 12 h. Experiment repeated three times. Scale bar, 200 μm. (b) Quantification of GTP-bound RhoA together with total cell lysates was assessed by western blot after Rhotekin pull-down assay. Cells were treated with DMSO (NT) or with GGTI-298 1 μM for 12 h. Experiment repeated three times.
Supplementary Figure 6 Activation of the mevalonate pathway increases YAP/TAZ activity.
(a) qPCR analysis in MDA-MB-231 to measure SREBPs transcriptional activity. MDA-MB-231 cells were plated in medium with 10%FBS or 2%ULTROSER (US) for 48 h. SREBPs target genes (HMGCS1 and HMGCR) expression was analysed by qPCR. Error bars represent mean ± s.d., from n = 3 biological replicates. (b) Nuclear YAP/TAZ localization in MDA-MB-231 cells placed in medium with 10% FBS or 2% ULTROSER (US) for 24 h. Data are derived from n=3 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (c) qPCR in MDA-MB-231 cells placed in medium with 10% FBS or 2% ULTROSER (US) for 24 h. Error bars represent mean s.d., from n=4 biological replicates. (d) Nuclear YAP/TAZ localization in confluent MCF10A MII cells placed in medium with 5% Horse Serum (H.S.) or 2% ULTROSER (US) and treated with Cerivastatin (CER) 1 μM for 24 h. Data are derived from n = 4 independent experiments where at least 300 cells were scored. Error bars represent mean ± s.d. (e) Representative images relative to d. Scale bars, 15 μm. (f) qPCR analysis in MDA-MB-231 to measure SREBPs transcriptional activity. MDA-MB-231 cells were placed in medium with 2% ULTROSER and treated with DMSO (NT) or SREBPs inhibitor Fatostatin 40 μM for 48 h. SREBPs target genes (HMGCS1 and HMGCR) expression was analysed by qPCR. Error bars represent mean ± s.d., from n = 3 biological replicates. (g) Cleaved SREBP-2 (nSREBP-2) levels were detected after 48 h treatment with DMSO (NT) or SREBPs inhibitor Fatostatin (FT) 40 μM in cells cultured with 10%FBS or 2%ULTROSER (US). Representative blots are shown. Experiment repeated three times. *P < 0.05, **P < 0.01; Students t-test is used throughout.
Supplementary Figure 7 Mutant-p53 activates YAP/TAZ through the mevalonate pathway.
(a) qPCR analysis in MDA-MB-231 to measure SREBPs transcriptional activity after mutant p53 silencing. SREBPs target genes (HMGCS1 and HMGCR) expression was analysed by qPCR. Error bars represent mean ± s.d., from n = 3 biological replicates. (b) p53 levels relative to Fig. 6e (upper panel) and 6i (bottom panel). Representative blots are shown. Experiment repeated three times. (c) Immunofluorescence images shown in Fig. 6f, here presented with their nuclear staining (Hoechst) and zoom. Scale bars, 15 μm. (d) Contingency table frequencies of samples classified as having high or low levels of Mevalonate Pathway signature, of YAP/TAZ signature and of mutant p53 ten genes signature. The association among high/low levels of Mevalonate signature, YAP/TAZ, and ten genes resulted statistically significant (Pearson’s Chi-squared Test, p < 10−16). n = 657 tumor samples. All error bars are s.d. (*P < 0.05, **P < 0.01; Students t-test is used throughout).
Supplementary information
Supplementary Information
Supplementary Information (PDF 1957 kb)
Rights and permissions
About this article
Cite this article
Sorrentino, G., Ruggeri, N., Specchia, V. et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol 16, 357–366 (2014). https://doi.org/10.1038/ncb2936
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2936
This article is cited by
-
Role of cell rearrangement and related signaling pathways in the dynamic process of tip cell selection
Cell Communication and Signaling (2024)
-
The LKB1–TSSK1B axis controls YAP phosphorylation to regulate the Hippo–YAP pathway
Cell Death & Disease (2024)
-
Cardiac fibroblasts and mechanosensation in heart development, health and disease
Nature Reviews Cardiology (2023)
-
Statins improve endothelial function via suppression of epigenetic-driven EndMT
Nature Cardiovascular Research (2023)
-
Verteporfin-induced proteotoxicity impairs cell homeostasis and survival in neuroblastoma subtypes independent of YAP/TAZ expression
Scientific Reports (2023)