Abstract
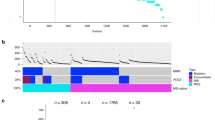
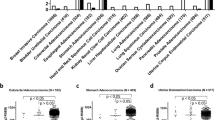
We report somatic mutations of RNF43 in over 18% of colorectal adenocarcinomas and endometrial carcinomas. RNF43 encodes an E3 ubiquitin ligase that negatively regulates Wnt signaling. Truncating mutations of RNF43 are more prevalent in microsatellite-unstable tumors and show mutual exclusivity with inactivating APC mutations in colorectal adenocarcinomas. These results indicate that RNF43 is one of the most commonly mutated genes in colorectal and endometrial cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Koo, B.K. et al. Nature 488, 665–669 (2012).
Jiang, X. et al. Proc. Natl. Acad. Sci. USA 110, 12649–12654 (2013).
Wu, J. et al. Proc. Natl. Acad. Sci. USA 108, 21188–21193 (2011).
Jiao, Y. et al. J. Pathol. 232, 428–435 (2014).
Hodis, E. et al. Cell 150, 251–263 (2012).
Cancer Genome Atlas Network. Nature 490, 61–70 (2012).
Stransky, N. et al. Science 333, 1157–1160 (2011).
Clevers, H. & Nusse, R. Cell 149, 1192–1205 (2012).
Cancer Genome Atlas Network. Nature 487, 330–337 (2012).
Seshagiri, S. et al. Nature 488, 660–664 (2012).
Liao, X. et al. N. Engl. J. Med. 367, 1596–1606 (2012).
Cancer Genome Atlas Research Network. Nature 497, 67–73 (2013).
Wang, K. et al. Nat. Genet. 43, 1219–1223 (2011).
Zang, Z.J. et al. Nat. Genet. 44, 570–574 (2012).
Wang, K. et al. Nat. Genet. 46, 573–582 (2014).
Cancer Genome Atlas Research Network. Nature 513, 202–209 (2014).
Van Allen, E.M. et al. Nat. Med. 20, 682–688 (2014).
Cibulskis, K. et al. Nat. Biotechnol. 31, 213–219 (2013).
Robinson, J.T. et al. Nat. Biotechnol. 29, 24–26 (2011).
Kim, D. & Salzberg, S.L. Genome Biol. 12, R72 (2011).
Acknowledgements
We thank S. Bahl, E. Nickerson and S. Chauvin for project management and A. Regev for help with computing resources, as well as A. Sivachenko, F. Huang and P. Tamayo for helpful discussions. This work was supported by the Dana-Farber Cancer Institute Leadership Council, the 2014 Colon Cancer Alliance–American Association for Cancer Research Fellowship for Biomarker Research, grant 14-40-40-GIAN, the Perry S. Levy Endowed Fellowship (M.G.), award T32GM007753 from the National Institute of General Medical Sciences, the Herchel Smith Fellowship (E.H.) and the Agrusa Fund for Colorectal Cancer Research (C.S.F.), as well as National Human Genome Research Institute grant U54HG003067 (E.S.L. and S.B.G.) and National Cancer Institute grants, K07CA190673 (R.N.), P01CA87969, UM1CA167552 and R01CA151993 (S.O.), R01CA118553 and R01CA168141 (C.S.F.) and P50CA127003 (M.G., W.C.H., L.A.G. and C.S.F.).
Author information
Authors and Affiliations
Contributions
M.G., E.H., W.C.H., G.G., S.B.G., E.S.L., S.O., C.S.F. and L.A.G. designed research. M.G., E.H., X.J.M., M.Y., J.R., K.C., G.S., M.S.L., Z.R.Q., R.N. and E.M.V.A. performed research. G.G., S.B.G., E.S.L., S.O. and C.S.F. contributed new reagents and analytic tools. M.G., E.H., X.J.M., E.M.V.A. and L.A.G. analyzed data. M.G., E.H. and L.A.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
L.A.G. is a consultant for an equity holder in Foundation Medicine. L.A.G. is also a consultant to Novartis, Millenium/Takeda and Boehringer Ingelheim and is a recipient of a grant from Novartis.
Integrated supplementary information
Supplementary Figure 1 Manual review of RNF43 p.Gly659fs mutations using Integrated Genomics Viewer and validation by Sanger sequencing.
Representative Integrated Genomics Viewer screenshots and Sanger sequencing chromatograms for RNF43 p.Gly659fs-mutated tumor (a,c) and matched normal (b,d) tissue.
Supplementary information
Supplementary Text and Figures
Manual review of RNF43 G659fs mutations using Integrated Genomics Viewer and validation by Sanger sequencing. (PDF 262 kb)
Supplementary Tables 1–10
Supplementary Tables 1–10 (XLSX 35911 kb)
Rights and permissions
About this article
Cite this article
Giannakis, M., Hodis, E., Jasmine Mu, X. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 46, 1264–1266 (2014). https://doi.org/10.1038/ng.3127
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3127
This article is cited by
-
New insights in ubiquitin-dependent Wnt receptor regulation in tumorigenesis
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Inherited BRCA1 and RNF43 pathogenic variants in a familial colorectal cancer type X family
Familial Cancer (2024)
-
Microsatellite instability assessment is instrumental for Predictive, Preventive and Personalised Medicine: status quo and outlook
EPMA Journal (2023)
-
Clinicopathological and molecular characteristics of RSPO fusion-positive colorectal cancer
British Journal of Cancer (2022)
-
RNF43 G659fs is an oncogenic colorectal cancer mutation and sensitizes tumor cells to PI3K/mTOR inhibition
Nature Communications (2022)