Abstract
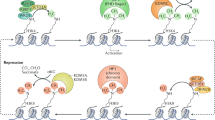
DNA methylation inhibits gene expression in animal cells, probably by affecting chromatin structure. Biochemical studies suggest that this process may be mediated by methyl-specific binding proteins that recruit enzymatic machinery capable of locally altering histone modification1. To test whether DNA methylation actually has a role in the assembly of chromatin during normal development, we used cell transfection and a transgene construct genetically programmed to be either methylated or unmethylated in all cell types of the mouse2. Chromatin immunoprecipitation (ChIP) analysis shows that the presence of DNA methylation brings about the deacetylation of histone H4 and methylation of Lys9 of histone H3 (H3 Lys9) and prevents methylation of Lys4 of histone H3 (H3 Lys4), thus generating a structure identical to that of methylated sequences in the genome. These results indicate that the methylation pattern established in early embryogenesis is profoundly important in setting up the structural profile of the genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Razin, A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J. 17, 4905–4908 (1998).
Siegfried, Z. et al. DNA methylation represses transcription in vivo. Nat. Genet. 22, 203–206 (1999).
Kafri, T. et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germline. Genes Dev. 6, 705–714 (1992).
Monk, M., Boubelik, M. & Lehnert, S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99, 371–382 (1987).
Brandeis, M. et al. Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438 (1994).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Litt, M.D., Simpson, M., Gaszner, M., Allis, C.D. & Felsenfeld, G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293, 2453–2455 (2001).
Wolffe, A.P. Histone H1. Int. J. Biochem. Cell Biol. 29, 1463–1466 (1997).
Wolffe, A.P., Khochbin, S. & Dimitrov, S. What do linker histones do in chromatin? Bioessays 19, 249–255 (1997).
Renz, M. Preferential and cooperative binding of histone I to chromosomal mammalian DNA. Proc. Natl. Acad. Sci. USA 72, 733–736 (1975).
Misteli, T., Gunjan, A., Hock, R., Bustin, M. & Brown, D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 (2000).
Cameron, E.E., Bachman, K.E., Myohanen, S., Herman, J.G. & Baylin, S.B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21, 103–107 (1999).
Nan, X. et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998).
Jones, P.L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 (1998).
Fuks, F. et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. (2002).
Wang, H. et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8, 1207–1217 (2001).
Briggs, S.D. et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 (2001).
Zegerman, P., Canas, B., Pappin, D. & Kouzarides, T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 277, 11621–11624 (2002).
Nishioka, K. et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16, 479–489 (2002).
Lallemand, Y., Luria, V., Haffner-Krausz, R. & Lonai, P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site specific recombinase. Transgenic Res. 7, 105–112 (1998).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
Eden, S., Hashimshony, T., Keshet, I., Thorne, A.W. & Cedar, H. DNA methylation models histone acetylation. Nature 394, 842–843 (1998).
Bustin, M. Preparation and application of immunological probes for nucleosomes. Methods Enzymol. 170, 214–251 (1989).
Hebbes, T.R., Clayton, A.L., Thorne, A.W. & Crane-Robinson, C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 13, 1823–1830 (1994).
Acknowledgements
We thank T. Jenuwein for his gift of branched methylated H3 Lys9 antibody. This research was supported by grants from the US National Institutes of Health, the Israel Cancer Research Fund and the Arthur and Rochelle Belfer Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hashimshony, T., Zhang, J., Keshet, I. et al. The role of DNA methylation in setting up chromatin structure during development. Nat Genet 34, 187–192 (2003). https://doi.org/10.1038/ng1158
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1158
This article is cited by
-
DNA demethylation and tri-methylation of H3K4 at the TACSTD2 promoter are complementary players for TROP2 regulation in colorectal cancer cells
Scientific Reports (2024)
-
Recent advancements in identification and detection of saliva as forensic evidence: a review
Egyptian Journal of Forensic Sciences (2023)
-
Purified regenerating retinal neurons reveal regulatory role of DNA methylation-mediated Na+/K+-ATPase in murine axon regeneration
Communications Biology (2023)
-
Epigenetic genes and epilepsy — emerging mechanisms and clinical applications
Nature Reviews Neurology (2022)
-
5-Azacytidine increases tanshinone production in Salvia miltiorrhiza hairy roots through epigenetic modulation
Scientific Reports (2022)