Abstract
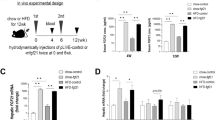
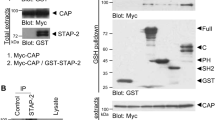
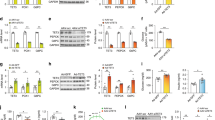
Using an expression cloning strategy, we have identified TFE3, a basic helix-loop-helix protein, as a transactivator of metabolic genes that are regulated through an E-box in their promoters. Adenovirus-mediated expression of TFE3 in hepatocytes in culture and in vivo strongly activated expression of IRS-2 and Akt and enhanced phosphorylation of insulin-signaling kinases such as Akt, glycogen synthase kinase 3β and p70S6 kinase. TFE3 also induced hexokinase II (HK2) and insulin-induced gene 1 (INSIG1). These changes led to metabolic consequences, such as activation of glycogen and protein synthesis, but not lipogenesis, in liver. Collectively, plasma glucose levels were markedly reduced both in normal mice and in different mouse models of diabetes, including streptozotocin-treated, db/db and KK mice. Promoter analyses showed that IRS2, HK2 and INSIG1 are direct targets of TFE3. Activation of insulin signals in both insulin depletion and resistance suggests that TFE3 could be a therapeutic target for diabetes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
White, M.F. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 182, 3–11 (1998).
Virkamaki, A., Ueki, K. & Kahn, C.R. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Invest. 103, 931–943 (1999).
Withers, D.J. et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391, 900–904 (1998).
Kubota, N. et al. Disruption of insulin receptor substrate 2 causes type 2 diabetes because of liver insulin resistance and lack of compensatory beta-cell hyperplasia. Diabetes 49, 1880–1889 (2000).
Rother, K.I. et al. Evidence that IRS-2 phosphorylation is required for insulin action in hepatocytes. J. Biol. Chem. 273, 17491–17497 (1998).
Kido, Y. et al. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Invest. 105, 199–205 (2000).
Rawson, R.B. The SREBP pathway–insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4, 631–640 (2003).
Shimano, H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 40, 439–452 (2001).
Horton, J.D., Goldstein, J.L. & Brown, M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Osborne, T.F. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275, 32379–32382 (2000).
Matsuzaka, T. et al. Insulin-independent induction of sterol regulatory element-binding protein-1c expression in the livers of streptozotocin-treated mice. Diabetes 53, 560–569 (2004).
Kerouz, N.J., Horsch, D., Pons, S. & Kahn, C.R. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J. Clin. Invest. 100, 3164–3172 (1997).
Shimomura, I. et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell 6, 77–86 (2000).
Yahagi, N. et al. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lep(ob)/Lep(ob) mice. J. Biol. Chem. 277, 19353–19357 (2002).
Ide, T. et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol. 6, 351–357 (2004).
Vallet, V.S. et al. Glucose-dependent liver gene expression in upstream stimulatory factor 2 −/− mice. J. Biol. Chem. 272, 21944–21949 (1997).
Casado, M., Vallet, V.S., Kahn, A. & Vaulont, S. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J. Biol. Chem. 274, 2009–2013 (1999).
Riu, E., Bosch, F. & Valera, A. Prevention of diabetic alterations in transgenic mice overexpressing Myc in the liver. Proc. Natl. Acad. Sci. USA 93, 2198–2202 (1996).
Shimano, H. et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 274, 35832–35839 (1999).
Uyeda, K., Yamashita, H. & Kawaguchi, T. Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem. Pharmacol. 63, 2075–2080 (2002).
Beckmann, H., Su, L.K. & Kadesch, T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 4, 167–179 (1990).
Giangrande, P.H., Hallstrom, T.C., Tunyaplin, C., Calame, K. & Nevins, J.R. Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23, 3707–3720 (2003).
Grinberg, A.V. & Kerppola, T. Both Max and TFE3 cooperate with Smad proteins to bind the plasminogen activator inhibitor-1 promoter, but they have opposite effects on transcriptional activity. J. Biol. Chem. 278, 11227–11236 (2003).
Feinman, R. et al. PU.1 and an HLH family member contribute to the myeloid-specific transcription of the Fc gamma RIIIA promoter. EMBO J. 13, 3852–3860 (1994).
Zhao, G.Q., Zhao, Q., Zhou, X., Mattei, M.G. & de Crombrugghe, B. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol. Cell. Biol. 13, 4505–4512 (1993).
Osawa, H., Sutherland, C., Robey, R.B., Printz, R.L. & Granner, D.K. Analysis of the signaling pathway involved in the regulation of hexokinase II gene transcription by insulin. J. Biol. Chem. 271, 16690–16694 (1996).
Yoon, J.C. et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 (2001).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 (2003).
Daitoku, H., Yamagata, K., Matsuzaki, H., Hatta, M. & Fukamizu, A. regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52, 642–649 (2003).
Riu, E. et al. Overexpression of c-myc in the liver prevents obesity and insulin resistance. FASEB J. 17, 1715–1717 (2003).
Yoshikawa, T. et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21, 2991–3000 (2001).
Amemiya-Kudo, M. et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43, 1220–1235 (2002).
Murakami, K. et al. A novel insulin sensitizer acts as a coligand for peroxisome proliferator-activated receptor-alpha (PPAR-alpha) and PPAR-gamma: effect of PPAR-alpha activation on abnormal lipid metabolism in liver of Zucker fatty rats. Diabetes 47, 1841–1847 (1998).
He, T.C. et al. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95, 2509–2514 (1998).
Heikkinen, S. et al. Mouse hexokinase II gene: structure, cDNA, promoter analysis, and expression pattern. Mamm. Genome 11, 91–96 (2000).
Peng, Y. et al. Cloning, human chromosomal assignment, and adipose and hepatic expression of the CL-6/INSIG1 gene. Genomics 43, 278–284 (1997).
Acknowledgements
This work was supported by grants-in-aid from the Ministry of Science, Education, Culture, and Technology of Japan (to Y.N., H.S., A.T. and N.Y.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
The multiple products of TFE3 are derived from different ATG sites. The Tcfe3 cDNA contains four potential start (ATG) sites. (PDF 309 kb)
Supplementary Fig. 2
The effects of dominant negative form of TFE3 (dnTFE3) on insulin signaling and glycogen synthesis in rat primary hepatocytes. (PDF 512 kb)
Supplementary Fig. 3
Dose-dependent effects of adenoviral TFE3 on hepatic gene expression as estimated by northern blotting. (PDF 320 kb)
Supplementary Fig. 4
HkII and INSIG1 promoters as targets of TFE3. (a) The mouse HkII promoter contains an E box which is responsible for induction of TFE3. (PDF 514 kb)
Supplementary Fig. 5
Physiological regulation of Irs2 expression in fasted and refed states, and effects of TFE3 over-expression on insulin-signaling. (PDF 351 kb)
Supplementary Table 1
Metabolic parameters in C57BL6 mice overexpressing GFP and wtTFE3. (PDF 296 kb)
Rights and permissions
About this article
Cite this article
Nakagawa, Y., Shimano, H., Yoshikawa, T. et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med 12, 107–113 (2006). https://doi.org/10.1038/nm1334
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1334
This article is cited by
-
Emerging roles of TFE3 in metabolic regulation
Cell Death Discovery (2023)
-
Emerging roles and regulation of MiT/TFE transcriptional factors
Cell Communication and Signaling (2018)
-
A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes
Nature Communications (2018)
-
CREB3L3 controls fatty acid oxidation and ketogenesis in synergy with PPARα
Scientific Reports (2016)
-
Translation control during prolonged mTORC1 inhibition mediated by 4E-BP3
Nature Communications (2016)