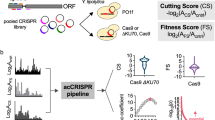
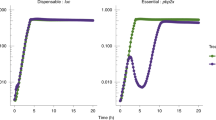
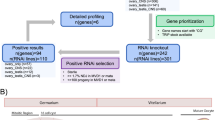
Abstract
This protocol describes the various steps and considerations involved in planning and carrying out RNA interference (RNAi) genome-wide screens in cultured Drosophila cells. We focus largely on the procedures that have been modified as a result of our experience over the past 3 years and of our better understanding of the underlying technology. Specifically, our protocol offers a set of suggestions and considerations for screen optimization and a step-by-step description of the procedures successfully used at the Drosophila RNAi Screening Center for screen implementation, data collection and analysis to identify potential hits. In addition, this protocol briefly covers postscreen analysis approaches that are often needed to finalize the hit list. Depending on the scope of the screen and subsequent analysis and validation involved, the full protocol can take anywhere from 3 months to 2 years to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Adams, M.D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000).
Hannon, G.J. RNA interference. Nature 418, 244–251 (2002).
Dykxhoorn, D.M., Novina, C.D. & Sharp, P.A. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4, 457–467 (2003).
Tomari, Y. & Zamore, P.D. Perspective: machines for RNAi. Genes Dev. 19, 517–529 (2005).
Moffat, J. & Sabatini, D.M. Building mammalian signalling pathways with RNAi screens. Nat. Rev. Mol. Cell Biol. 7, 177–187 (2006).
Clemens, J.C. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97, 6499–6503 (2000).
Hammond, S.M., Bernstein, E., Beach, D. & Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 (2000).
Lum, L. et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039–2045 (2003).
Kiger, A.A. et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2, 27 (2003).
Foley, E. & O'Farrell, P.H. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2, e203 (2004).
Boutros, M. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004).
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R.A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002).
Feske, S. et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179–185 (2006).
Vig, M. et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220–1223 (2006).
Zhang, S.L. et al. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA 103, 9357–9362 (2006).
Perrimon, N. & Mathey-Prevot, B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175, 7–16 (2007).
Kennerdell, J.R. & Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95, 1017–1026 (1998).
Kennerdell, J.R. & Carthew, R.W. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol 18, 896–898 (2000).
Kim, Y.O., Park, S.J., Balaban, R.S., Nirenberg, M. & Kim, Y. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc. Natl. Acad. Sci. USA 101, 159–164 (2004).
Koizumi, K. et al. RNA interference screen to identify genes required for Drosophila embryonic nervous system development. Proc. Natl. Acad. Sci. USA 104, 5626–5631 (2007).
Ma, Y., Creanga, A., Lum, L. & Beachy, P.A. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359–363 (2006).
Kulkarni, M.M. et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 3, 833–838 (2006).
Echeverri, C.J. et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3, 777–779 (2006).
Perrimon, N. & Mathey-Prevot, B. Matter arising: off targets and genome scale RNAi screens in Drosophila. Fly 1, e1–e5 (2007).
Armknecht, S. et al. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392, 55–73 (2005).
Friedman, A. & Perrimon, N. High-throughput approaches to dissecting MAPK signaling pathways. Methods 40, 262–271 (2006).
Kallio, J. et al. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 7, 811–819 (2005).
Björklund, M. et al. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439, 1009–1013 (2006).
Boutros, M., Bras, L. & Huber, W. Analysis of cell-based RNAi screens. Genome Biol. 7, R66 (2006).
Zhang, X.D. et al. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics 7, 299–309 (2006).
Fuchs, F. & Boutros, M. Cellular phenotyping by RNAi. Brief Funct. Genomic. Proteomic. 5, 52–56 (2006).
Mathey-Prevot, B. & Perrimon, N. Regulatory RNAs: Cold Spring Harbor Symposia on Quantitative Biology Vol. LXXI. (eds. Stillman B. & Stewart D.) (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2007).
Cully, M.J. & Leevers, S.J. RNA interference pinpoints regulators of cell size and the cell cycle. Genome Biol. 7, 219 (2006).
Echeverri, C.J. & Perrimon, N. High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 7, 373–384 (2006).
Hild, M. et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 5, R3 (2003).
Malo, N., Hanley, J.A., Cerquozzi, S., Pelletier, J. & Nadon, R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175 (2006).
Makarenkov, V. et al. An efficient method for the detection and elimination of systematic error in high-throughput screening. Bioinformatics 23, 1648–1657 (2007).
Gentleman, R., Carey, V.J., Huber, W., Irizarri, R. & Dudoit, S. Bioinformatics and Computational Biology Solutions using R and Bioconductor (Springer, New York, 2005).
Sims, D., Bursteinas, B., Gao, Q., Zvelebil, M. & Baum, B. FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34, D479–D483 (2006).
Flockhart, I. et al. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34, D489–D494 (2006).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Carpenter, A.E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Bera, A.K. & Jarque, C.M. Efficient tests for normality, homoscedasticity and serial independence of regression residuals. Econ. Lett. 6, 255–259 (1980).
Friedman, A. & Perrimon, N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature 444, 230–234 (2006).
Wheeler, D.L. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35, D5–D12 (2007).
O'Brien, K.P., Remm, M. & Sonnhammer, E.L. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33, D476–D480 (2005).
Hirsh, A.E. & Fraser, H.B. Protein dispensability and rate of evolution. Nature 411, 1046–1049 (2001).
Jordan, I.K., Rogozin, I.B., Wolf, Y.I. & Koonin, E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12, 962–968 (2002).
Crosby, M.A., Goodman, J.L., Strelets, V.B., Zhang, P. & Gelbart, W.M. FlyBase: genomes by the dozen. Nucleic Acids Res. 35, D486–D491 (2007).
Grumbling, G. & Strelets, V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34, D484–488 (2006).
Pacifico, S. et al. A database and tool, IM Browser, for exploring and integrating emerging gene and protein interaction data for Drosophila. BMC Bioinformatics 7, 195 (2006).
DasGupta, R., Kaykas, A., Moon, R.T. & Perrimon, N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826–833 (2005).
Schneider, I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353–365 (1972).
Yanagawa, S., Lee, J.S. & Ishimoto, A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 273, 32353–32359 (1998).
Echalier, G. & Ohanessian, A. [Isolation, in tissue culture, of Drosophila melanogaster cell lines]. C. R. Acad. Sci. Hebd. Seances. Acad. Sci. D 268, 1771–1773 (1969).
Haars, R., Zentgraf, H., Gateff, E. & Bautz, F.A. Evidence for endogenous reovirus-like particles in a tissue culture cell line from Drosophila melanogaster. Virology 101, 124–130 (1980).
Ui, K. et al. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. In Vitro Cell. Dev. Biol. Anim. 30A, 209–216 (1994).
Ui, K., Ueda, R. & Miyake, T. Cell lines from imaginal discs of Drosophila melanogaster. In Vitro Cell. Dev. Biol. 23, 707–711 (1987).
Acknowledgements
We thank all members of the Perrimon laboratory as well as all past and present DRSC screeners for sharing their experience with us. None of the protocols described here would have been possible without their contributions. Work at the DRSC is supported by grant R01 GM067761 from the NIGMS. N.P. is an investigator of the Howard Hughes Medical School.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ramadan, N., Flockhart, I., Booker, M. et al. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc 2, 2245–2264 (2007). https://doi.org/10.1038/nprot.2007.250
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.250
This article is cited by
-
NetREX-CF integrates incomplete transcription factor data with gene expression to reconstruct gene regulatory networks
Communications Biology (2022)
-
TRiC/CCT chaperonins are essential for organ growth by interacting with insulin/TOR signaling in Drosophila
Oncogene (2019)
-
RNA interference-mediated knockdown of the hydroxyacid-oxoacid transhydrogenase gene decreases thiamethoxam resistance in adults of the whitefly Bemisia tabaci
Scientific Reports (2017)
-
ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation
Nature Cell Biology (2015)
-
Adapting human pluripotent stem cells to high-throughput and high-content screening
Nature Protocols (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.