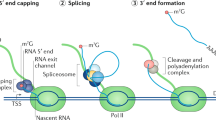
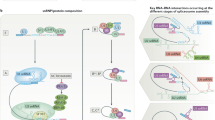
Key Points
-
Bacterial fermentation products with cytostatic and antitumour activity target components of the basal precursor mRNA (pre-mRNA) splicing machinery.
-
At least some of these drugs achieve their effects by interfering with mechanisms that ensure proper recognition of 3′ splice site sequences by spliceosomal U2 small nuclear ribonucleoprotein (snRNP) particles.
-
Rather than global inhibition of the splicing process, cytostatic drug concentrations induce changes in alternative pre-mRNA splicing and downregulation of genes important for cell cycle and tumour progression.
-
The drugs display significantly more potent effects on cancer cells than on normal cells and retain their activity on multidrug-resistant cells.
-
The drug target splicing factor 3B subunit 1 (SF3B1) is a protein component of U2 snRNP that is frequently mutated in myelodysplasias with ring sideroblasts and in chronic lymphocytic leukaemia.
Abstract
Several bacterial fermentation products and their synthetic derivatives display antitumour activities and bind tightly to components of the spliceosome, which is the complex molecular machinery involved in the removal of introns from mRNA precursors in eukaryotic cells. The drugs alter gene expression, including alternative splicing, of genes that are important for cancer progression. A flurry of recent reports has revealed that genes encoding splicing factors, including the drug target splicing factor 3B subunit 1 (SF3B1), are among the most highly mutated in various haematological malignancies such as chronic lymphocytic leukaemia and myelodysplastic syndromes. These observations highlight the role of splicing factors in cancer and suggest that an understanding of the molecular effects of drugs targeting these proteins could open new perspectives for studies of the spliceosome and its role in cancer progression, and for the development of novel antitumour therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Moore, M. J. & Proudfoot, N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009).
Wahl, M. C., Will, C. L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Will, C. L. & Luhrmann, R. Spliceosome structure and function. Cold Spring. Harb. Perspect. Biol. 3, a003707 (2011).
Blencowe, B. J. Alternative splicing: new insights from global analyses. Cell 126, 37–47 (2006).
Nilsen, T. W. & Graveley, B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010).
Cooper, T. A., Wan, L. & Dreyfuss, G. RNA and disease. Cell 136, 777–793 (2009).
Kalsotra, A. & Cooper, T. A. Functional consequences of developmentally regulated alternative splicing. Nature Rev. Genet. 12, 715–729 (2011).
David, C. J. & Manley, J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010).
Kaida, D., Schneider-Poetsch, T. & Yoshida, M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 103, 1611–1616 (2012).
Pio, R. & Montuenga, L. M. Alternative splicing in lung cancer. J. Thorac. Oncol. 4, 674–678 (2009).
Harper, S. J. & Bates, D. O. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nature Rev. Cancer 8, 880–887 (2008).
Cheng, J. et al. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 263, 1759–1762 (1994).
Christofk, H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008).
Jeyaraj, S., O'Brien, D. M. & Chandler, D. S. MDM2 and MDM4 splicing: an integral part of the cancer spliceome. Front. Biosci. 14, 2647–2656 (2009).
Kole, R., Krainer, A. R. & Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nature Rev. Drug Discov. 11, 125–140 (2012).
Hua, Y. et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010).
Hua, Y. et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126 (2011).
Nakajima, H. et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. (Tokyo) 49, 1204–1211 (1996).
Nakajima, H. et al. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. (Tokyo) 49, 1196–1203 (1996).
Nakajima, H., Takase, S., Terano, H. & Tanaka, H. New antitumor substances, FR901463, FR901464 and FR901465. III. Structures of FR901463, FR901464 and FR901465. J. Antibiot. (Tokyo) 50, 96–99 (1997).
Mizui, Y. et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J. Antibiot. (Tokyo) 57, 188–196 (2004).
Sakai, T., Asai, N., Okuda, A., Kawamura, N. & Mizui, Y. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. II. Physico-chemical properties and structure elucidation. J. Antibiot. (Tokyo) 57, 180–187 (2004).
Sakai, T. et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. I. Taxonomy, fermentation, isolation and screening. J. Antibiot. (Tokyo) 57, 173–179 (2004).
Sakai, Y. et al. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. II. The effects on cell cycle progression and gene expression. J. Antibiot. (Tokyo) 55, 863–872 (2002).
Sakai, Y. et al. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. (Tokyo) 55, 855–862 (2002).
Albert, B. J. et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol. Cancer Ther. 8, 2308–2318 (2009).
Kaida, D. et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nature Chem. Biol. 3, 576–583 (2007).
Kotake, Y. et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nature Chem. Biol. 3, 570–575 (2007). References 27 and 28 describe the discovery of two families of drugs identified by their cytostatic effects (causing arrest in G1 and G2/M phases of the cell cycle) as well as their antitumour properties in animal models. Biochemical experiments identified SF3B, a subcomplex of spliceosomal U2 snRNP particles, as a target of the drugs. Biochemical assays and experiments in cell lines demonstrate inhibition of pre-mRNA splicing by these compounds.
Fan, L., Lagisetti, C., Edwards, C. C., Webb, T. R. & Potter, P. M. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 6, 582–589 (2011). This paper describes the total synthesis, in gram quantities, of sudemycins, which are drug analogues that possess the putative common pharmacophore between the SSA-related compound FR901464 and pladienolide B. Sudemycins are potent regulators of alternative splicing. They are significantly more active on tumour cells than on normal cells, enhancing their therapeutic potential in oncology.
Lagisetti, C. et al. Synthetic mRNA splicing modulator compounds with in vivo antitumor activity. J. Med. Chem. 52, 6979–6990 (2009).
Lagisetti, C. et al. Antitumor compounds based on a natural product consensus pharmacophore. J. Med. Chem. 51, 6220–6224 (2008).
Webb, T. R., Joyner, A. S. & Potter, P. M. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov. Today 3 Aug 2012 (doi:10.1016/j.drudis.2012.07.013).
Hasegawa, M. et al. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem. Biol. 6, 229–233 (2011).
Yokoi, A. et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 278, 4870–4880 (2011).
Corrionero, A., Minana, B. & Valcarcel, J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 25, 445–459 (2011). Biochemical experiments show that SSA destabilizes 3′ splice site recognition by U2 snRNP, preventing interactions of its target SF3B1 with the pre-mRNA and allowing base pairing of U2 snRNA to decoy, unproductive sites. Alternative 3′ splice sites can be differentially sensitive to the drug depending on the presence of such decoy sites, which leads to changes in alternative splicing in genes important for cell cycle progression, thus providing a potential molecular mechanism for the antitumour effects of the drug.
Roybal, G. A. & Jurica, M. S. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 38, 6664–6672 (2010).
Schmidt, U. et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J. Cell Biol. 193, 819–829 (2011).
Abu Dayyeh, B. K., Quan, T. K., Castro, M. & Ruby, S. W. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J. Biol. Chem. 277, 20221–20233 (2002).
Perriman, R. & Ares, M. Jr. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell 38, 416–427 (2010).
Folco, E. G., Coil, K. E. & Reed, R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 25, 440–444 (2011). Biochemical experiments demonstrate that E7107, a pladienolide antitumour drug, modulates the equilibrium between two alternative conformations of U2 snRNA, which results in defective 3′ splice site recognition by U2 snRNP. The results provide a novel mechanism for splicing modulation by small molecules.
An, M. & Henion, P. D. The zebrafish sf3b1b460 mutant reveals differential requirements for the sf3b1 pre-mRNA processing gene during neural crest development. Int. J. Dev. Biol. 56, 223–237 (2012).
Martins, S. B. et al. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nature Struct. Mol. Biol. 18, 1115–1123 (2011).
Brody, Y. et al. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 9, e1000573 (2011).
Furumai, R. et al. Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Sci. 101, 2483–2489 (2010).
Allende-Vega, N. et al. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 20 Feb 2012 (doi:10.1038/onc.2012.38).
Hahn, C. N. & Scott, H. S. Spliceosome mutations in hematopoietic malignancies. Nature Genet. 44, 9–10 (2012).
Edery, P. et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332, 240–243 (2011).
He, H. et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332, 238–240 (2011).
Golas, M. M., Sander, B., Will, C. L., Luhrmann, R. & Stark, H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol. Cell 17, 869–883 (2005).
Massiello, A., Roesser, J. R. & Chalfant, C. E. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20, 1680–1682 (2006).
Friend, K., Lovejoy, A. F. & Steitz, J. A. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol. Cell 28, 240–252 (2007).
Kyburz, A., Friedlein, A., Langen, H. & Keller, W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell 23, 195–205 (2006).
Di Giammartino, D. C., Nishida, K. & Manley, J. L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011).
Neilson, J. R. & Sandberg, R. Heterogeneity in mammalian RNA 3′ end formation. Exp. Cell Res. 316, 1357–1364 (2010).
Isono, K., Mizutani-Koseki, Y., Komori, T., Schmidt-Zachmann, M. S. & Koseki, H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 19, 536–541 (2005).
Abdel-Wahab, O. & Levine, R. The spliceosome as an indicted conspirator in myeloid malignancies. Cancer Cell 20, 420–423 (2011).
Damm, F. et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 119, 3211–3218 (2012).
Visconte, V., Makishima, H., Maciejewski, J. P. & Tiu, R. V. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia 15 May 2012 (doi:10.1038/leu.2012.130).
Lindsley, R. C. & Ebert, B. L. Molecular pathophysiology of myelodysplastic syndromes. Annu. Rev. Pathol. 28 Aug 2012 (doi:10.1146/annurev-pathol-011811-132436).
Murati, A. et al. Myeloid malignancies: mutations, models and management. BMC Cancer 12, 304 (2012).
Damm, F., Nguyen-Khac, F., Fontenay, M. & Bernard, O. A. Spliceosome and other novel mutations in chronic lymphocytic leukemia and myeloid malignancies. Leukemia 26, 2027–2031 (2012).
Makishima, H. et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210 (2012).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). Whole-exome sequencing of samples from MDSs reveal frequent mutations in genes encoding splicing factors, particularly proteins involved in 3′ splice site recognition, such as U2AF35 and SF3B1. The mutations are mutually exclusive and correlate with abnormal RNA splicing. The results represent a compelling argument for splicing alterations as contributors to the pathogenesis of myeloid neoplasms.
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Hirabayashi, S. et al. Spliceosomal gene aberrations are rare, coexist with oncogenic mutations, and are unlikely to exert a driver effect in childhood MDS and JMML. Blood 119, e96–e99 (2012).
Matsuda, K. et al. Long-term haematological improvement after non-intensive or no chemotherapy in juvenile myelomonocytic leukaemia and poor correlation with adult myelodysplasia spliceosome-related mutations. Br. J. Haematol. 157, 647–650 (2012).
Lasho, T. L. et al. SF3B1 mutations in primary myelofibrosis: clinical, histopathology and genetic correlates among 155 patients. Leukemia 26, 1135–1137 (2012).
Visconte, V. et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 26, 542–545 (2012).
Malcovati, L. et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 118, 6239–6246 (2011).
Cui, R. et al. Clinical importance of SF3B1 mutations in Chinese with myelodysplastic syndromes with ring sideroblasts. Leuk. Res. 36, 1428–1433 (2012).
Patnaik, M. M. et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood 119, 569–572 (2012).
Jeromin, S. et al. High frequencies of SF3B1 and JAK2 mutations in refractory anemia with ring sideroblasts associated with marked thrombocytosis strengthen the assignment to the category of myelodysplastic/myeloproliferative neoplasms. Haematologica 28 Aug 2012 (doi:10.3324/haematol.2012.072538).
Visconte, V. et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood 23 Jul 2012 (doi:10.1182/blood-2012-05-430876). RSs are the most conspicuous pathological feature correlating with mutations in SF3B1 in myelodysplasias. The authors show that pharmacological inhibition of SF3B1 induces RSs in healthy bone marrow cells, and that bone marrow aspirates from SF3B1 heterozygous knockout mice also contain RSs. These results demonstrate that the RS phenotype is caused by loss of function of SF3B1, possibly associated with splicing defects.
Thol, F. et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 119, 3578–3584 (2012).
Quesada, V. et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature Genet. 44, 47–52 (2012).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011).
Rossi, D. et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 118, 6904–6908 (2011). References 75–77 describe the results of whole-exome sequencing of CLL samples, revealing frequent mutations in the gene encoding the splicing factor SF3B1 that correlate with faster disease progression, resistance to fludarabine and poor overall survival. A striking pattern of recurrent amino acid changes was observed, clustering in the HEAT repeats domain. Alterations in RNA processing associated with the presence of the mutations were found, offering a potential mechanism to understand the contribution of RNA processing to CLL progression.
Quesada, V., Ramsay, A. J. & Lopez-Otin, C. Chronic lymphocytic leukemia with SF3B1 mutation. N. Engl. J. Med. 366, 2530 (2012).
Pleasance, E. D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007).
Ellis, M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
Groves, M. R. & Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 (1999).
Xing, Y. et al. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 127, 341–353 (2006).
Golas, M. M., Sander, B., Will, C. L., Luhrmann, R. & Stark, H. Molecular architecture of the multiprotein splicing factor SF3b. Science 300, 980–984 (2003).
Graubert, T. A. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature Genet. 44, 53–57 (2012).
Wu, S. J. et al. The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood 29 Aug 2012 (doi:10.1182/blood-2012-02-412296).
Ogle, J. M. & Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 (2005).
Berg, M. G. et al. A quantitative high throughput in vitro splicing assay identifies inhibitors of spliceosome catalysis. Mol. Cell. Biol. 32, 1271–1283 (2012).
O'Brien, K., Matlin, A. J., Lowell, A. M. & Moore, M. J. The biflavonoid isoginkgetin is a general inhibitor of pre-mRNA splicing. J. Biol. Chem. 283, 33147–33154 (2008).
Shkreta, L. et al. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol. Cancer Ther. 7, 1398–1409 (2008).
Roca, X. et al. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 26, 1098–1109 (2012).
Smith, D. J., Konarska, M. M. & Query, C. C. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol. Cell 34, 333–343 (2009).
Gozani, O., Potashkin, J. & Reed, R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18, 4752–4760 (1998).
Warkocki, Z. et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nature Struct. Mol. Biol. 16, 1237–1243 (2009).
Lardelli, R. M., Thompson, J. X., Yates, J. R. & Stevens, S. W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16, 516–528 (2010).
Wang, C. et al. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12, 1409–1414 (1998).
Egecioglu, D. E. & Chanfreau, G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA 17, 383–389 (2011).
Wang, Z. & Burge, C. B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813 (2008).
Chen, M. & Manley, J. L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Rev. Mol. Cell Biol. 10, 741–754 (2009).
Witten, J. T. & Ule, J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 27, 89–97 (2011).
Barash, Y. et al. Deciphering the splicing code. Nature 465, 53–59 (2010).
Heyd, F. & Lynch, K. W. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem. Sci. 36, 397–404 (2011).
Kornblihtt, A. R. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 623, 175–189 (2007).
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R. & Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011).
Zaharieva, E., Chipman, K. & Soller, M. Alternative splicing interference by xenobiotics. Toxicology 296, 1–12 (2012).
Yoon, S. O., Shin, S., Lee, H. J., Chun, H. K. & Chung, A. S. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol. Cancer Ther. 5, 2666–2675 (2006).
Stoilov, P., Lin, C. H., Damoiseaux, R., Nikolic, J. & Black, D. L. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl Acad. Sci. USA 105, 11218–11223 (2008).
Anderson, E. S. et al. The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. RNA 18, 1041–1049 (2012).
Soret, J. et al. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc. Natl Acad. Sci. USA 102, 8764–8769 (2005).
Ghigna, C. et al. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol. 7, 495–503 (2010).
Mermoud, J. E., Cohen, P. & Lamond, A. I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 20, 5263–5269 (1992).
Zhang, M. L., Lorson, C. L., Androphy, E. J. & Zhou, J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther. 8, 1532–1538 (2001).
Zhang, Z. et al. Synthesis and characterization of pseudocantharidins, novel phosphatase modulators that promote the inclusion of exon 7 into the SMN (survival of motoneuron) pre-mRNA. J. Biol. Chem. 286, 10126–10136 (2011).
Pilch, B. et al. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 61, 6876–6884 (2001).
Muraki, M. et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 279, 24246–24254 (2004).
Nishida, A. et al. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nature Commun. 2, 308 (2011).
Fukuhara, T. et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl Acad. Sci. USA 103, 11329–11333 (2006).
Chang, J. G. et al. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS ONE 6, e18643 (2011).
Chang, W. H. et al. Amiloride modulates alternative splicing in leukemic cells and resensitizes Bcr-AblT315I mutant cells to imatinib. Cancer Res. 71, 383–392 (2011).
Younis, I. et al. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol. Cell. Biol. 30, 1718–1728 (2010).
Ghosh, A. K. & Anderson, D. D. Enantioselective total synthesis of pladienolide B: a potent spliceosome inhibitor. Org. Lett. 14, 4730–4733 (2012).
Osman, S. et al. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chemistry 17, 895–904 (2011).
Acknowledgements
We thank P. Cironi, C. López-Otín, P. Potter, V. Quesada, J. Vilardell, T. Webb and members of our laboratory for discussions and comments on the manuscript, and three anonymous reviewers for useful suggestions. Work in our group has been supported by Fundación Botín, Consolider RNAREG, Ministerio de Economía y Competitividad, AGAUR, AICR and EURASNET.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Bonnal, S., Vigevani, L. & Valcárcel, J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11, 847–859 (2012). https://doi.org/10.1038/nrd3823
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd3823
This article is cited by
-
Splicing factor-mediated regulation patterns reveals biological characteristics and aid in predicting prognosis in acute myeloid leukemia
Journal of Translational Medicine (2023)
-
Comprehensive characterization of 11 prognostic alternative splicing events in ovarian cancer interacted with the immune microenvironment
Scientific Reports (2022)
-
Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors
Nature Communications (2021)
-
Overlapping roles of spliceosomal components SF3B1 and PHF5A in rice splicing regulation
Communications Biology (2021)
-
Alternative splicing and cancer: a systematic review
Signal Transduction and Targeted Therapy (2021)