Key Points
-
The cloning of the complementary DNAs and genes for the insulin and insulin-like growth factor 1 (IGF1) receptors, as well as tertiary-structure predictions, have provided valuable insights into the overall domain organization of the receptors.
-
No crystal structure of the insulin- or IGF1-receptor complexes with their ligands is yet available, but crystal structures of the large domain 1 (L1)–Cys-rich (CR)–L2 amino-terminal fragment of the IGF1 receptor, and of the insulin- and IFG1-receptor tyrosine-kinase domains both in the inactive and activated conformation, are available.
-
Single-molecule electron-microscopic imaging of the insulin receptor has given some indications of the overall organization of the extracellular receptor domains, although there have been variable results and a tendency in some cases to over-interpret low-resolution data.
-
Knowledge of the structure of several receptor tyrosine-kinase domains has led to attempts to screen for, or design, mimetics or inhibitors, with some degree of success. Agonists or antagonists that target the ligand-binding sites might have a greater chance to be selective, hence the importance of understanding the nature of the ligand-binding mechanism.
-
Mapping the ligand-binding sites on the insulin and IGF1 receptors by receptor crosslinking with photoreactve ligands, by examining the binding selectivity of chimeric insulin–IGF1 receptors, by alanine-scanning mutagenesis of receptor domains and by reconstitution of minimized receptor constructs with low or high affinity, has provided a wealth of information on the binding epitopes.
-
Likewise, mapping of the residues on the insulin molecule that are involved in receptor binding has progressed, and has revealed the existence of a second binding surface in addition to the so-called 'classical' binding surface. The information on IGF1 and IGF2 is more fragmentary.
-
Alternative crosslinking models that explain the complex ligand-binding kinetics of the insulin and IGF1 receptors (including negative cooperativity) are discussed.
-
Various strategies for designing agonists or antagonists of a dimerizing receptor are discussed, building on the experience acquired with the erythropoietin receptor.
Abstract
Type 2 diabetes mellitus — in which the body produces insufficient amounts of insulin or the insulin that is produced does not function properly to control blood glucose — is an increasingly common disorder. Prospective clinical studies have proven the benefits of tighter glucose control in reducing the frequency and severity of complications of the disease, leading to the advocation of earlier and more aggressive use of insulin therapy. Given the reluctance of patients with type 2 diabetes to inject themselves with insulin, orally active insulin mimetics would be a major therapeutic advance. Here, we discuss recent progress in understanding the structure–function relationships of the insulin and insulin-like growth factor 1 (IGF1) receptors, their mechanism of activation and their implications for the design of insulin-receptor agonists for diabetes therapy and IGF1-receptor antagonists for cancer therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Amos, A. F., McCarty, D. J. & Zimmet, P. The rising global burden of diabetes and its complications: estimates and projections to the year 2010. Diabetic Med. 14 (Suppl. 5), S1–S85 (1997).
Bilous, R. W. & Alberti, K. G. M. M. in New Antidiabetic Drugs (eds Bailey, C. J. & Flat, P. R.) 19–32 (Smith-Gordon and Nishimura, London, United Kingdom and Niigata–Shi, Japan, 1990).
Yudkin, J. S. Therapeutic targets for type 2 diabetes post-UKPDS. J. R. Coll. Phys. Lond. 34, 254–256 (2000).
Standl, E. Insulin analogues — state of the art. Horm. Res. 57 (Suppl. 1), 40–45 (2002).
Owens, D. R. New horizons — alternative routes for insulin therapy. Nature Rev. Drug Discov. 1, 529–540 (2002).
De Meyts, P. Insulin and insulin-like growth factors: the paradox of signaling specificity. Growth Horm. IGF Res. 12, 81–83 (2002).
Adams, T. E., Epa, V. C., Garrett, T. P. & Ward, C. W. Structure and function of the type 1 insulin-like growth factor receptor. Cel. Mol. Life Sci. 57, 1050–1093 (2000)A thorough review of the structure–function relationships of the IGF1 and insulin receptors.
Blume, A. J., Beasley, J. & Goldstein, N. I. The use of peptides in diogenesis: a novel approach to drug discovery and phenomics. Biopolymers 55, 347–356 (2000).
Khandwala, H. M., McCutcheon, I. E., Flyvbjerg, A. & Friend, K. E. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endo. Rev. 21, 215–244 (2000).
Ebina, Y. et al. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell 40, 747–758 (1985).
Ullrich, A. et al. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 313, 756–761 (1985).References 10 and 11 report the cloning of the insulin receptor cDNA (the two groups happened to clone one of the alternative isoforms; see reference 19).
Whittaker, J. et al. High-level expression of human insulin receptor cDNA in mouse NIH 3T3 cells. Proc. Natl Acad. Sci. USA 84, 5237–5241 (1987).
Ullrich, A. et al. Insulin-like growth factor-I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 5, 2503–2512 (1986).Cloning of the IGF1-receptor cDNA.
Seino, S., Seino, M., Nishi, S. & Bell, G. I. Structure of the human insulin receptor gene and characterization of its promoter. Proc. Natl Acad. Sci. USA 86, 114–118 (1989).
Abbott, A. M., Bueno, R., Pedrini, M. T., Murray, J. M. & Smith, R. J. Insulin-like growth factor I receptor gene structure. J. Biol. Chem. 267, 10759–10763 (1992).References 14 and 15 report the sequencing of the insulin- and IGF1-receptor genes and their relationship to the modular structure of the receptors.
Hubbard, S. R. & Till, J. H. Protein tyrosine kinase structure and function. Ann. Rev. Biochem. 69, 373–398 (2000).
Heldin, C. H. & Ostman, A. Ligand-induced dimerization of growth factor receptors: variations on the theme. Cytokine Growth Factor Rev. 7, 3–10 (1996).
Bass, J., Chiu, G., Argon, Y. & Steiner, D. F. Folding of insulin receptor monomers is facilitated by the molecular chaperones calnexin and calreticulin and impaired by rapid dimerization. J. Cell Biol. 141, 637–646 (1998).
Seino, S. & Bell, G. I. Alternative splicing of human insulin receptor messenger RNA. Biochem. Biophys. Res. Comm. 159, 312–316 (1989).
Soos, M., Field, C. E. & Siddle, K. Purified hybrid insulin/insulin-like growth factor-I receptors bind insulin-like growth factor-I, but not insulin, with high affinity. Biochem. J. 290, 419–426 (1993).
Pandini, G. et al. Insulin/IGF1 hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J. Biol. Chem. 2002 Jul 22 (doi: 10.1074/jbc.M202766200).
Bajaj, M., Waterfield, M. D., Schlessinger, J., Taylor, W. R. & Blundell, T. L. On the tertiary structure of the extracellular domains of the epidermal growth factor and insulin receptors. Biochim. Biophys. Acta 916, 220–226 (1987).An insightful attempt at predicting the tertiary structure of the amino-terminal domain of the insulin, IGF1 and EGF receptors that proved to be largely correct.
Ward, C. W., Hoyne, P. A. & Flegg, R. H. Insulin and epidermal growth factor receptors contain the cysteine repeat motif found in the tumor necrosis factor receptor. Proteins 22, 141–153 (1995).
Marino-Buslje, C., Mizuguchi, K., Siddle, K. & Blundell, T. L. A third fibronectin type III domain in the extracellular region of the insulin receptor family. FEBS Lett. 441, 331–336 (1998).
Mulhern, T. D., Booker, G. W. & Cosgrove, L. A third fibronectin-type-III domain in the insulin-family receptors. Trends Biol. Sci. 23, 465–466 (1998).
Ward, C. W. Members of the insulin receptor family contain three fibronectin type III domains. Growth Factors 16, 315–322 (1999).
Garrett, T. P. et al. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature 394, 395–399 (1998).This study determined the three-dimensional structure of the amino-terminal domain of the IGF1 receptor, and confirmed the predictions of references 22 and 23 . This fragment does not bind the ligand.
Kristensen, C., Wiberg, F. C., Schäffer, L. & Andersen, A. S. Expression and characterization of a 70-kDa fragment of the insulin receptor that binds insulin. Minimizing ligand binding domain of the insulin receptor. J. Biol. Chem. 273, 17780–17786 (1998).The first demonstration that addition of a 12-residue carboxy-terminal peptide confers binding affinity to the domain that was crystallized in reference 27.
Molina, L., Marino-Buslje, C., Quinn, D. R. & Siddle, K. Structural domains of the insulin receptor and IGF receptor required for dimerisation and ligand binding. FEBS Lett. 467, 226–230 (2000).
Hubbard, S. R., Wei, L., Ellis, L. & Hendrickson, W. A. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372, 746–754 (1994).
Hubbard, S. R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16, 5572–5581 (1997).
Favelyukis, S., Till, J. H., Hubbard, S. R. & Miller, W. T. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nature Struct. Biol. 8, 1058–1063 (2001).
Pautsch, A. et al. Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation. Structure 9, 955–965 (2001).References 30–33 report the three-dimensional structures of the inactive and activated insulin- and IGF1-receptor kinases.
Schaefer, E. M., Erickson, H. P., Federwisch, M., Wollmer, A. & Ellis, L. Structural organization of the human insulin receptor ectodomain. J. Biol. Chem. 267, 23393–23402 (1992).
Tulloch, P. A. et al. Single-molecule imaging of human insulin receptor ectodomain and its Fab complexes. J. Struct. Biol. 125, 11–18 (1999).An electron micrograph structure of the insulin-receptor ectodomain is validated by the use of monoclonal antibodies.
Woldin, C. N., Hing, F. S., Lee, J., Pilch, P. F. & Shipley, G. G. Structural studies of the detergent-solubilized and vesicle-reconstituted insulin receptor. J. Biol. Chem. 274, 34981–34992 (1999).
Christiansen, K., Tranum-Jensen, J., Carlsen, J. & Vinten, J. A model for the quaternary structure of human placental insulin receptor deduced from electron microscopy. Proc. Natl Acad. Sci. USA 88, 249–252 (1991).
Tranum-Jensen, J., Christiansen, K., Carlsen, J., Brenzel, G. & Vinten, J. Membrane topology of insulin receptors reconstituted into lipid vesicles. J. Membr. Biol. 140, 215–223 (1994).
Luo, R. Z., Beniac, D. R., Fernandes, A., Yip, C. C. & Ottensmeyer, F. P. Quaternary structure of the insulin–insulin receptor complex. Science 285, 1077–1080 (1999).
Ottensmeyer, F. P., Beniac, D. R., Luo, R. Z. & Yip, C. C. Mechanism of transmembrane signaling: insulin binding and the insulin receptor. Biochemistry 39, 12103–12112 (2000).References 39 and 40 are an attempt at fitting three-dimensional structures of insulin-receptor domains in cryoelectron microscopic images; details might have to be taken with a pinch of salt.
Slade, A., Luh, J., Ho, S. & Yip, C. M. Single molecule imaging of supported planar lipid bilayer-reconstituted human insulin receptors by in situ scanning probe microscopy. J. Struct. Biol. 137, 283–291 (2002).
Saltiel, A. R. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 104, 517–529 (2001).
Frick, W., Bauer, A., Bauer, J., Wied, S. & Muller, G. Structure–activity relationship of synthetic phosphoinositolglycans mimicking metabolic insulin action. Biochemistry 37, 13421–13436 (1998).
Elchebly, M. et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 (1999).
Clement, S. et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 409, 92–97 (2001).
Zhang, B. et al. Discovery of a small molecule insulin mimetic with antidiabetic activity in mice. Science 284, 974–977 (1999).Not likely to end up in clinical trials, but establishes the proof-of-concept for direct insulin-receptor-kinase activation.
Air, E. L. et al. Small molecule insulin mimetics reduce food intake and body weight and prevent development of obesity. Nature Med. 8, 179–183 (2002).
Manchem, V. P. et al. A novel small molecule that directly sensitizes the insulin receptor in vitro and in vivo. Diabetes 50, 824–830 (2001).
Levitzki, A. & Gazit, A. Tyrosine kinase inhibition: an approach to drug development. Science 267, 1782–1788 (1995).
Mendel, D. B. et al. Development of SU5416, a selective small molecule inhibitor of VEGF receptor tyrosine kinase activity, as an anti-angiogenesis agent. Anticancer Drug Des. 15, 29–41 (2000).
Rewcastle, G. W. et al. Tyrosine kinase inhibitors. 14. Structure–activity relationships for methylamino-substituted derivatives of 4-[(3-bromophenyl)amino]-6-(methylamino)-pyrido[3,4-d]pyrimidine (PD 158780), a potent and specific inhibitor of the tyrosine kinase activity of receptors for the EGF family of growth factors. J. Med. Chem. 41, 742–751 (1998).
Mohammadi, M. et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 17, 5896–5904 (1998).
Bilder, G. et al. Restenosis following angioplasty in the swine coronary artery is inhibited by an orally active PDGF-receptor tyrosine kinase inhibitor, RPR101511A. Circulation 99, 3292–3299 (1999).
Parang, K. et al. Mechanism-based design of a protein kinase inhibitor. Nature Struct. Biol. 8, 37–41 (2001).
De Meyts, P., Roth, J., Neville, J., Gavin, J. R. & Lesniak, M. A. Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem. Biophys. Res. Comm. 55, 154–161 (1973).
De Meyts, P., Van Obberghen, E., Roth, J., Brandenburg, D. & Wollmer, A. Mapping of the residues of the receptor binding region of insulin responsible for the negative cooperativity. Nature 273, 504–509 (1978).
De Meyts, P. The structural basis of insulin and insulin-like growth factor-I (IGF1) receptor binding and negative cooperativity, and its relevance to mitogenic versus metabolic signaling. Diabetologia 37 (Suppl. 2), S135–S148 (1994).References 55–57 retrace the history of the concept of negative cooperativity at the insulin receptor from initial discovery, to description of the cooperative site, to discussion of plausible models that integrate known kinetic data.
Christoffersen, C. T. et al. Negative cooperativity in the insulin-like growth factor-I (IGF1) receptor and a chimeric IGF1/insulin receptor. Endocrinology 135, 472–475 (1994).
Zhang, B. & Roth, R. A. A region of the insulin receptor important for ligand binding (residues 450–601) is recognized by patients autoimmune antibodies and inhibitory monoclonal antibodies. Proc. Natl Acad. Sci. USA 88, 9858–9862 (1991).
Kadowaki, H. et al. Mutagenesis of lysine 460 in the human insulin receptor. Effects upon receptor recycling and cooperative interactions among binding sites. J. Biol. Chem. 265, 21285–21296 (1990).
Kjeldsen, T. et al. The ligand specificities of the insulin receptor and the insulin-like growth factor-I receptor reside in different regions of a common binding site. Proc. Natl Acad. Sci. USA 88, 4404–4408 (1991).
Andersen, A. S. et al. Identification of determinants that confer ligand specificity on the insulin receptor. J. Biol. Chem. 267, 13681–13686 (1992).
Schumacher, R., Mosthaf, L., Schlessinger, J., Brandenburg, D. & Ullrich, A. Insulin and insulin-like growth factor-I binding specificity is determined by distinct regions of their cognate receptors. J. Biol. Chem. 266, 19288–19295 (1991).References 54–56 provide clues to the structural basis for ligand selectivity of the insulin and IGF1 receptors.
Tavare, J. M. & Siddle, K. Mutational analysis of insulin receptor function: consensus and controversy. Biochim. Biophys. Acta 1178, 21–39 (1993).
Hoyne, P. A., Elleman, T. C., Adams, T. E., Richards, K. M. & Ward, C. W. Properties of an insulin receptor with an IGF-1 receptor loop exchange in the cysteine-rich region. FEBS Lett. 469, 57–60 (2000).
Williams, P. F., Mynarcik, D. C., Qin, Y. G. & Whittaker, J. Mapping of an NH2-terminal ligand binding site of the insulin receptor by alanine scanning mutagenesis. J. Biol. Chem 270, 1–5 (1995).The first alanine-scanning mutagenesis of the insulin receptor.
De Meyts, P. et al. Identification of a ligand-binding region of the human insulin receptor encoded by the second exon of the gene. Molecular Endocrinology 4, 409–416 (1990).
Mynarcik, D. C., Yu, G. Q. & Whittaker, J. Alanine-scanning mutagenesis of a C-terminal ligand binding domain of the insulin receptor α subunit. J. Biol. Chem. 271, 2439–2442 (1996).
Mynarcik, D. C., Williams, P. F., Schffer, L., Yu, G. Q. & Whittaker, J. Analog binding properties of insulin receptor mutants. Identification of amino acids interacting with the COOH terminus of the B chain of the insulin molecule. J. Biol. Chem 272, 2077–2081 (1997).
Taylor, S. I. et al. Mutations in the insulin receptor gene. Endocr. Rev. 13, 566–595 (1992).
Kadowaki, T., Kadowaki, H., Accili, D. & Taylor, S. I. Substitution of lysine for asparagine at position 15 in the α-subunit of the human insulin receptor. A mutation that impairs transport of receptors to the cell surface and decreases the affinity of insulin binding. J. Biol. Chem. 265, 19143–19150 (1990).
Rouard, M. et al. Congenital insulin resistance associated with a conformational alteration in a conserved beta-sheet in the insulin receptor L1 domain. J. Biol. Chem. 274, 18487–18491 (1999).
Longo, N., Langley, S. D., Griffin, L. D. & Elsas, L. J. Activation of glucose transport by a natural mutation in the human insulin receptor. Proc. Natl Acad. Sci. USA 90, 60–64 (1993).
Grønskov, K., Vissing, H., Shymko, R. M., Tornqvist, H. & De Meyts, P. Mutation of arginine 86 to proline in the insulin receptor alpha subunit causes lack of transport of the receptor to the plasma membrane, loss of binding affinity and a constitutively activated tyrosine kinase in transfected cells. Biochem. Biophys. Res. Comm. 192, 905–911 (1993).
Nakae, J., Morioka, H., Ohtsuka, E. & Fujieda, K. Replacements of leucine 87 in human insulin receptor alter affinity for insulin. J. Biol. Chem. 270, 22017–22022 (1995).
Liu, R. et al. Deletion of lysine 121 creates a temperature-sensitive alteration in insulin binding by the insulin receptor. J. Biol. Chem. 270, 476–482 (1995).
Hamer, I. et al. An arginine to cysteine 252 mutation in insulin receptors from a patient with severe insulin resistance inhibits receptor internalisation but preserves signalling events. Diabetologia 45, 657–667 (2002).
Roach, P. et al. A novel human insulin receptor gene mutation uniquely inhibits insulin binding without impairing posttranslational processing. Diabetes 43, 1096–1102 (1994).
Whittaker, J. & Mynarcik, D. C. Phenotype of the Ser-323 to Leu mutation of the insulin receptor is isoform dependent. Diabetes 47 (Suppl. 1), 264 (1998).
Whittaker, J. et al. Alanine scanning mutagenesis of a type 1 insulin-like growth factor receptor ligand binding site. J. Biol. Chem. 276, 43980–43986 (2001).
Schäffer, L. A model for insulin binding to the insulin receptor. Eur. J. Biochem. 221, 1127–1132 (1994).The first plausible model of the insulin-receptor binding mechanism that integrates and explains all known kinetic parameters.
Shoelson, S. E., Lee, J., Lynch, C. S., Backer, J. M. & Pilch, P. F. BpaB25 insulins. Photoactivatable analogues that quantitatively crosslink, radiolabel, and activate the insulin receptor. J. Biol. Chem. 268, 4085–4091 (1993).
Wang, C. C. et al. Negative and positive site–site interactions, and their modulation by pH, insulin analogs and monoclonal antibodies, are preserved in the purified insulin receptor. Proc. Natl Acad. Sci. USA 85, 8400–8404 (1988).
Markussen, J. M., Halstrom, J., Wiberg, F. & Schffer, L. Immobilized insulin for high capacity affinity chromatography of insulin receptors. J. Biol. Chem. 266, 18814–18818 (1991).
Yip, C. C. & Jack, E. Insulin receptors are bivalent as demonstrated by photoaffinity labeling. J. Biol. Chem. 267, 13131–13134 (1992).
Sweet, L. J., Morrison, B. D. & Pessin, J. Isolation of functional a-b heterodimers from the purified human placental α2–β2 heterotetrameric insulin receptor concept. Structural basis for high affinity ligand binding. J. Biol. Chem. 262, 6939–6942 (1987).
Boni-Schnetzler, M., Scott, W., Waugh, S. M., Di Bella, E. & Pilch, P. The insulin receptor. Structural basis for high affinity ligand binding. J. Biol. Chem. 262, 8395–8401 (1987).
Bass, J., Kurose, T., Pashmforoush, M. & Steiner, D. F. Fusion of insulin receptor ectodomains to immunoglobulin constant domains reproduces high-affinity insulin binding in vitro. J. Biol. Chem. 271, 19367–19375 (1996).
Hoyne, P. A. et al. High affinity insulin binding by soluble insulin receptor extracellular domain fused to a leucine zipper. FEBS Lett. 479, 15–18 (2000).
Whittaker, J., Garcia, P., Yu, G. Q. & Mynarcik, D. C. Transmembrane domain interactions are necessary for negative cooperativity of the insulin receptor. Mol. Endocrinol. 8, 1521–1527 (1994).
Florke, R. R. et al. Hormone-triggered conformational changes within the insulin-receptor ectodomain: requirement for transmembrane anchors. Biochem. J. 360, 189–198 (2001).
Baker, E. et al. The structure of 2Zn pig insulin at 1. 5 Å resolution. Phil. Trans. R. Soc. Lond. B 19, 369–456 (1988).The 'bible' of insulin structure.
Blundell. T. L., Dodson, G. G., Hodgkin, D. C. & Mercola, D. A. Insulin: the structure in the crystal and its reflection in chemistry and biology. Adv. Prot. Chem. 26, 279–402 (1972).The old testament.
Pullen, R. A. et al. Receptor-binding region of insulin. Nature 259, 369–373 (1976).The first description of the 'classical' insulin-binding surface.
Hua, X. H., Shoelson, S. E., Kochoyan, M. & Weiss, M. Receptor binding redefined by a structural switch in a mutant human insulin. Nature 354, 238–241 (1991).
Ludvigsen, S., Olsen, H. B. & Kaarsholm, N. C. A structural switch in a mutant insulin exposes key residues for receptor binding. J. Mol. Biol. 279, 1–7 (1998).References 95 and 96 describe a structural switch whereby the carboxy terminus of the B-chain of insulin moves away on receptor binding to uncover underlying A2–A3 residues, but disagrees on the extent of switching. Stay tuned.
Kristensen, C. et al. Alanine scanning mutagenesis of insulin. J. Biol. Chem. 272, 12978–12983 (1997).
Nakagawa, S. H. & Tager, H. S. Importance of aliphatic side-chain structure at positions 2 and 3 of the insulin A chain in insulin-receptor interactions. Biochemistry 31, 3204–3214 (1992).
Nakagawa, S. H., Tager, H. S. & Steiner, D. F. Mutational analysis of invariant valine B12 in insulin: implications for receptor binding. Biochemistry 39, 15826–15835 (2000).
van den Brande, J. L. in The Insulin-Like Growth Factors. Structure And Biological Functions (Oxford Univ. Press, Oxford, New York and Tokyo, 1992).
Brzozowski, A. M. et al. Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry 41, 9389–9397 (2002).
Vajdos, F. F. et al. Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions. Biochemistry 40, 11022–11029 (2001).References 101 and 102 provide crystal structures of IGF1, for which previously only NMR structures existed. There is a better definition of the carboxy-terminal peptide in reference 101.
Bayne, M. L. et al. The C region of human insulin-like growth factor (IGF) I is required for high affinity binding to the type I IGF receptor. J. Biol. Chem. 264, 11004–11008 (1988).
Gill, R. et al. Engineering the C-region of human insulin-like growth factor-1: implications for receptor binding. Protein Eng. 9, 1011–1019 (1996).
Kristensen, C. et al. A single-chain insulin-like growth factor I/insulin hybrid binds with high affinity to the insulin receptor. Biochem. J. 305, 981–986 (1995).
Chen, L. M., Yang, X. W. & Tang, J. G. Acidic residues on the N-terminus of proinsulin C-peptide are important for the folding of insulin precursor. J. Biochem. 131, 855–859 (2002).
de Vos, A. M., Ultsch, M. & Kossiakoff, A. A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255, 306–312 (1992).The first crystal structure of a ligand–receptor complex and a surprising stoichiometry.
Raff, M. in Physical Chemical Aspects Of Cell Surface Events In Cellular Regulation (eds DeLisi, C. & Blumenthal, R.) 116 (Elsevier North Holland, New York, Amsterdam, Oxford, 1979).“I am not suggesting insulin is aggregating with insulin in crosslinking. I am suggesting that insulin has more than one binding site for the receptor”. He was right.
Lee, J., O'Hare, T., Pilch, P. & Shoelson, S. E. Insulin receptor autophosphorylation occurs asymmetrically. J. Biol. Chem. 268, 4092–4098 (1993).
Yip, C. C. The insulin-binding domain of the insulin receptor is encoded by exons 2 and 3. J. Cell. Biochem. 48, 19–25 (1992).
Hammond, B. J., Tikerpae, J. & Smith, G. D. An evaluation of the crosslinking model for the interaction of insulin with its receptor. Am. J. Physiol. 272, E1136–E1144 (1997).
Yeh, J. I. et al. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 262, 186–201 (1996).A nice crystal structure and a good model for negative cooperativity at the insulin receptor.
Wedekind, F. et al. Hormone binding site of the insulin receptor: an analysis using photoaffinity-mediated avidin complexing. Biol. Chem. 370, 251–258 (1989).The first mapping of an insulin-receptor binding domain by photoaffinity crosslinking.
Kristensen, C., Andersen, A. S., Ostergaard, S., Hansen, P. H. & Brandt, J. Functional reconstitution of insulin receptor binding site from non-binding receptor fragments. J. Biol. Chem. 277, 18340–18345 (2002).
Schumacher, R. et al. Signaling-competent chimeras allow mapping of major insulin receptor binding domains determinants. J. Biol. Chem. 268, 1087–1094 (1993).
Fuh, G. et al. Rational design of potent antagonists to the human growth hormone receptor. Science 256, 1677–1680 (1992).A discussion of the important concept of antagonism at dimerizing receptors.
Okada, S. & Kopchick, J. J. Biological effects of growth hormone and its antagonist. Trends Mol. Med. 7, 126–132 (2001).
Boesen, T. P., Soni, B., Schwartz, T. & Halkier, T. Single-chain vascular endothelial growth factor variant with antagonist activity. J. Biol. Chem. 2002 Jul 31 (doi: 10.1074/jbc.M204107200).
Schlein, M. et al. Properties of small molecules affecting insulin receptor function. Biochemistry 40, 13520–13528 (2001).
Johnson, D. L. & Jolliffe, L. K. Erythropoietin mimetic peptides and the future. Nephrol. Dial. Transplant. 15, 1274–1277 (2000).
Clackson, T. & Wells, J. A. A hot spot of binding energy in a hormone–receptor interface. Science 267, 383–386 (1995).This paper discusses the concept of the 'functional epitope', another important concept from the group at Genentech.
Sidhu, S. S., Lowman, H. B., Cunningham, B. C. & Wells, J. A. Phage display for selection of novel binding peptides. Methods Enzymol. 328, 333–363 (2000).
Livnah, O. et al. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2. 8 Å Science 273, 464–471 (1996).
Wrighton, N. C. et al. Increased potency of an erythropoietin peptide mimetic through covalent dimerization. Nature Biotechnol. 15, 1261–1265 (1997).
Johnson, D. L. et al. Amino-terminal dimerization of an erythropoietin mimetic peptide results in increased erythropoietic activity. Chem. Biol. 4, 939–950 (1997).
Naranda, T. et al. Activation of erythropoietin receptor through a novel extracellular binding site. Endocrinology 143, 2293–2302 (2002).
Christopoulos, A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nature Rev. Drug Discov. 1, 198–210 (2002).
Qureshi, S. A. et al. Mimicry of erythropoietin by a nonpeptide molecule. Proc. Natl Acad. Sci. USA 96, 12156–12161 (1999).
Erlanson, D. A. et al. Site-directed ligand discovery. Proc. Natl Acad. Sci. USA 97, 9367–9372 (2000).
Budin, N., Ahmed, S., Majeux, N. & Caflisch, A. An evolutionary approach for structure-based design of natural and non-natural peptidic ligands. Comb. Chem. High Throughput Screen. 4, 661–673 (2001).
Fairbrother, W. J. et al. Novel peptides selected to bind vascular endothelial growth factor target the receptor-binding site. Biochemistry 37, 17754–17764 (1998).
Pillutla, R. C. et al. Peptides identify the critical hotspots involved in the biological activation of the insulin receptor. J. Biol. Chem. 277, 22590–22594 (2002).
Bursavich, M. G. & Rich, D. H. Designing non-peptide peptidomimetics in the 21st century: inhibitors targeting conformational ensembles. J. Med. Chem. 45, 541–558 (2002).
Ward, C. W. & Garrett, T. P. The relationship between the L1 and L2 domains of the insulin and epidermal growth factor receptors and leucine-rich repeat modules. BMC Bioinformatics 2, 4 (2001).
Schäffer, L. & Ljungqvist, L. Identification of a disulfide bridge connecting the alpha-subunits of the extracellular domain of the insulin receptor. Biochem. Biophys. Res. Comm. 189, 650–653 (1992).
Sparrow, L. G. et al. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 272, 29460–29467 (1997).
Lu, K. & Guidotti, G. Identification of the cysteine residues involved in the class I disulfide bonds of the human insulin receptor: properties of insulin receptor monomers. Mol. Biol. Cell 7, 679–691 (1996).
Cheatham, B. & Kahn, C. R. Cysteine 647 in the insulin receptor is required for normal covalent interaction between α- and β-subunits and signal transduction. J. Biol. Chem. 267, 7108–7115 (1992).
Yamaguchi, Y. et al. Functional properties of two naturally occuring isoforms of the human insulin receptor in chinese hamster ovary cells. Endocrinology 129, 2058–2066 (1991).
Gu, J. L. & De Meyts, P. in Retrospect and Prospect of Protein Research (eds Zai-Ping, L., Zi-Xian, L. & You-Shang, Z.) 120–125 (World Scientific Publishing, Singapore, 1991).
McClain, D. A. Different ligand affinities of the two human insulin receptor splice variants are reflected in parallel changes in sensitivity for insulin action. Mol. Endocrinol. 5, 734–739 (1991).
Frasca, F. et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol. Cell. Biol. 19, 3278–3288 (1999).
Pashmforoush, M., Yoshimasa, Y. & Steiner, D. F. Exon 11 enhances insulin binding affinity and tyrosine kinase activity of the human insulin proreceptor. J. Biol. Chem. 269, 32639–32648 (1994).
Marino-Buslje, C., Martin-Martinez, M., Mizuguchi, K., Siddle, K. & Blundell, T. L. The insulin receptor: from protein sequence to structure. Biochem. Soc. Trans. 27, 715–726 (1999).
Ward, C. W., Garrett, T. P., McKern, N. M. & Lawrence, L. J. Structure of the insulin receptor family: unexpected relationships with other proteins. Today. Life Sci. 11, 26–32 (1999).
Fabry, M. et al. Detection of a new hormone contact site within the insulin receptor ectodomain by the use of a novel photoreactive insulin. J. Biol. Chem. 267, 8950–8956 (1992).
Kurose, T. et al. Crosslinking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the α-subunit of the insulin receptor. J. Biol. Chem. 269, 29190–29197 (1994).A surprising crosslinking result that paved the way for interesting experiments (such as the ones in references 28, 29 and 114 ) that show the importance of a small carboxy-terminal domain for high-affinity binding.
Brandt, J., Andersen, A. S. & Kristensen, C. Dimeric fragment of the insulin receptor α-subunit binds insulin with full holoreceptor affinity. J. Biol. Chem. 276, 12378–12384 (2001).
Surinya, K. H. et al. Role of insulin receptor dimerization domains in ligand binding, cooperativity, and modulation by anti-receptor antibodies. J. Biol. Chem. 277, 16718–16725 (2002).References 148 and 149 provide an elegant deconvolution of the minimal-domain topography that is required for high-affinity insulin binding.
Waugh, S. M., DiBella, E. E. & Pilch, P. F. Isolation of a proteolitically derived domain of the insulin receptor containing the major site of crosslinking/binding. Biochemistry 28, 3448–3455 (1989).
Yip, C. C. et al. Localization of the insulin-binding site to the cysteine-rich region of the insulin receptor α-subunit. Biochem. Biophys. Res. Comm. 157, 321–329 (1988).
Cho, H.-S. & Leahy, D. J. Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297, 1330–1333 (2002).
Acknowledgements
We are grateful to J. Brandt and M. Krogsgaard Thomsen for critical review of the manuscript. The Receptor Biology Laboratory and the Hagedorn Research Institute are independent basic research components of Novo Nordisk A/S. The authors are also supported by grants from the Juvenile Diabetes Research Foundation International, the Danish Research Council through the Danish Center for Growth and Regeneration, and the Øresund region's Research and Development Committee (Øforsk).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
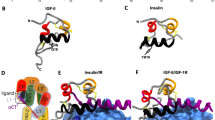
Animated figure 4| Insulin-like growth factor See legend below.
a | The three-dimensional structure of the large domain 1 (L1)–Cys-rich (CR)–L2 domain of the insulin-like growth factor 1 (IGF1) receptor determined by X-ray crystallography 17. An extended bi-lobed structure (40 × 48 × 105 Å) comprises the two globular L-domains with a new type of right-handed β-helix fold that flank the CR domain. They seem to be part of the leucine-rich-repeat superfamily 134. Although L1 (residues 1–150; green) contacts the CR domain (blue) along its length, there is minimal contact with L2 (residues 300–460; orange). The CR domain comprises an array of disulphide-linked modules that resemble those in the tumour-necrosis factor (TNF) receptor and laminin23. The different orientations of L1 and L2 relative to the CR domain are probably an artefact of crystal packing, and the position of L2 is probably more parallel to L1 in the native structure. The flexibility between the CR domain and L2 might be important for ligand binding 16. A cavity of ∼30 Å diameter occupies the centre of the molecule and represents a potential binding pocket, although this construct does not bind IGF1.The amino acids that have been determined by alanine-scanning mutagenesis 80 to be important for ligand binding are shown in yellow as van der Waals spheres (see main text for details). The three-dimensional structure of the IGF1 molecule is shown, based on the X-ray coordinates of Brzozowski and colleagues101. The amino acids that have been determined by site-directed mutagenesis to be important for receptor binding are shown in yellow as van der Waals spheres (reviewed in Ref. 7). As Arg36 and Arg37 are lacking in the structure, residues 35 and 38 have been highlighted instead to show their approximate location in the middle of the C-peptide domain. The backbone is shown in blue. b | The insulin-receptor L1–CR–L2 domain has been modelled on the corresponding domain coordinates of the IGF1-receptor using the programme SwissModel. The amino acids that have been determined by site-directed mutagenesis to be important for receptor binding66 are shown in yellow as van der Waals spheres. The three-dimensional structure of the insulin molecule is shown, based on the X-ray coordinates of Baker and colleagues92. The classical binding surface is shown in yellow as van der Waals spheres; Leu A13 and Leu B17 are shown in red. The backbone is shown in blue. Modelling was carried out using the DS ViewerLite 5.0 (Accelrys). (GIF 321 kb)
Related links
Related links
DATABASES
LocusLink
Medscape DrugInfo
OMIM
FURTHER INFORMATION
Glossary
- HYPOGLYCAEMIC AGENT
-
A drug that lowers blood glucose.
- TRANS-PHOSPHORYLATION
-
An enzymatic mechanism whereby one receptor-tyrosine-kinase domain transfers the γ-phosphate group of ATP (adenosine trisphosphate) to tyrosyl side chains of the second tyrosine-kinase domain of the same receptor (as opposed to the tyrosine-kinase domain phosphorylating itself, which would be cis-phosphorylation).
- SCATCHARD PLOT
-
A graphical method designed by George Scatchard in 1949 that consists of plotting the ratio of bound/free ligand against the concentration of bound ligand (as in figure 5a). In the case of simple (non-cooperative) binding, the graph is a straight line with a slope of −Ka, the affinity constant of the reaction.
- NEGATIVE COOPERATIVITY
-
A property of a multi-site-binding protein (for example an enzyme or a receptor) whereby the binding of one ligand molecule decreases the binding affinity of other ligand molecules to neighbouring binding sites.
- ALANINE SCANNING
-
The systematic replacement of the amino acids of a ligand or a binding protein by alanine (which just has a methyl group side chain) in order to evaluate the importance of the replaced side chains in the binding process.
- HOLORECEPTOR
-
The full-length receptor, as opposed to various minimized constructs, such as those shown in Box 2.
Rights and permissions
About this article
Cite this article
De Meyts, P., Whittaker, J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov 1, 769–783 (2002). https://doi.org/10.1038/nrd917
Issue Date:
DOI: https://doi.org/10.1038/nrd917
This article is cited by
-
Gene Characterization, Molecular Docking and Dynamic Simulations of Insulin-Like Growth Factor Receptor (IGF-1Ra) in Common Carp, Cyprinus carpio
Proceedings of the Zoological Society (2023)
-
Synergistic activation of the insulin receptor via two distinct sites
Nature Structural & Molecular Biology (2022)
-
Activation of the human insulin receptor by non-insulin-related peptides
Nature Communications (2022)
-
The role of insulin and incretin-based drugs in biliary tract cancer: epidemiological and experimental evidence
Discover Oncology (2022)
-
Increased O-GlcNAcylation promotes IGF-1 receptor/PhosphatidyI Inositol-3 kinase/Akt pathway in cervical cancer cells
Scientific Reports (2022)