Key Points
-
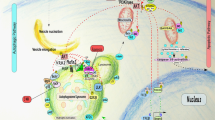
Autophagy and apoptosis constitute functionally distinct mechanisms for the turnover or destruction of cytoplasmic structures within cells and of cells within organisms, respectively.
-
Multiple stress-elicited signal transduction pathways can sequentially induce autophagy and apoptosis within the same cell. Elements of such pathways include p53, BH3 (BCL-2 homology 3)-only proteins, several kinases (including AKT, DAPK (death-associated protein kinase) and JNK (JUN N-terminal kinase)) and oncoproteins such as MYC and RAS.
-
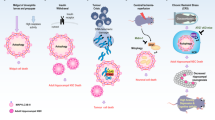
Autophagy increases the threshold of stress required for the induction of cell death by several mechanisms. These include the selective removal of damaged, potentially apoptosis-inducing mitochondria or that of other potentially lethal organelles, such as damaged zymogen granules in the exocrine pancreas. Autophagy can also lead to the selective elimination of pro-apoptotic signal transducers.
-
Apoptosis-associated caspase activation leads to the cleavage of various essential pro-autophagic proteins. Several of the resulting fragments acquire a new, pro-apoptotic function. This mechanism may accelerate the apoptotic demise of cells at its final stage.
-
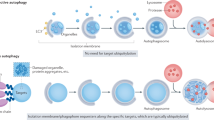
Cell death frequently occurs with (or is preceded by) autophagy, but it is rarely truly mediated by autophagy. In some cases, autophagic membranes or individual autophagy-relevant proteins facilitate the activation of apoptotic or necrotic pathways. There are also a few examples in which autophagy may be responsible for lethally dismantling cells. This cell death by autophagy is referred to as autophagic cell death (ACD).
-
Soluble products released by dying or dead cells can induce autophagy in neighbouring cells via the activation of pattern recognition receptors (PRRs). Pre-mortem autophagy is important for the production of chemotactic signals, which determine the removal of dead cells, inflammatory reactions and immune responses against dead-cell antigens.
-
Crosstalk between autophagy and apoptosis, as it occurs within the same cell or between distinct cells, has a cardinal role in pathophysiological processes, including those related to cancer, hormesis and ageing.
Abstract
Autophagy and apoptosis control the turnover of organelles and proteins within cells, and of cells within organisms, respectively, and many stress pathways sequentially elicit autophagy, and apoptosis within the same cell. Generally autophagy blocks the induction of apoptosis, and apoptosis-associated caspase activation shuts off the autophagic process. However, in special cases, autophagy or autophagy-relevant proteins may help to induce apoptosis or necrosis, and autophagy has been shown to degrade the cytoplasm excessively, leading to 'autophagic cell death'. The dialogue between autophagy and cell death pathways influences the normal clearance of dying cells, as well as immune recognition of dead cell antigens. Therefore, the disruption of the relationship between autophagy and apoptosis has important pathophysiological consequences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Maiuri, M. C., Zalckvar, E., Kimchi, A. & Kroemer, G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Rev. Mol. Cell Biol. 8, 741–752 (2007).
Yang, Z. & Klionsky, D. J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131 (2010).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Choi, A. M., Ryter, S. W. & Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 368, 1845–1846 (2013).
Thumm, M. et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280 (1994).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Yuan, J. & Kroemer, G. Alternative cell death mechanisms in development and beyond. Genes Dev. 24, 2592–2602 (2010).
Denton, D., Nicolson, S. & Kumar, S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 19, 87–95 (2012).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Weidberg, H., Shvets, E. & Elazar, Z. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 (2011).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010). References 11 and 12 enumerate the methods to quantify autophagy, as well as possible pitfalls in the interpretation of autophagy assays.
Galluzzi, L. et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107 (2009).
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Yu, L. et al. Regulation of an ATG7–beclin 1 program of autophagic cell death by caspase-8. Science 304, 1500–1502 (2004).
Shimizu, S. et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nature Cell Biol. 6, 1221–1228 (2004).
Boya, P. et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 25, 1025–1040 (2005). Provides the first evidence that autophagy inhibits apoptosis in mammalian cells.
Shen, H. M. & Codogno, P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy 7, 457–465 (2011).
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Tasdemir, E. et al. Regulation of autophagy by cytoplasmic p53. Nature Cell Biol. 10, 676–687 (2008).
Morselli, E. et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 10, 2763–2769 (2011).
Yu, X. et al. Inhibition of autophagy via p53-mediated disruption of ULK1 in a SCA7 polyglutamine disease model. J. Mol. Neurosci. 50, 586–599 (2013).
Vaseva, A. V. et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149, 1536–1548 (2012).
Galluzzi, L., Kepp, O. & Kroemer, G. Mitochondria: master regulators of danger signalling. Nature Rev. Mol. Cell Biol. 13, 780–788 (2012).
Youle, R. J. & Narendra, D. P. Mechanisms of mitophagy. Nature Rev. Mol. Cell Biol. 12, 9–14 (2011).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nature Rev. Mol. Cell Biol. 9, 402–412 (2008).
Budanov, A. V. & Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460 (2008).
Gao, W., Shen, Z., Shang, L. & Wang, X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 18, 1598–1607 (2011).
Kenzelmann Broz, D. et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016–1031 (2013). A complete catalogue of autophagy-relevant genes induced by p53.
He, Z. et al. p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene. Cell Death Differ. 20, 1415–1424 (2013).
Crighton, D. et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134 (2006).
Zhang, X. D., Qi, L., Wu, J. C. & Qin, Z. H. DRAM1 regulates autophagy flux through lysosomes. PloS ONE 8, e63245 (2013).
Laforge, M. et al. DRAM triggers lysosomal membrane permeabilization and cell death in CD4+T cells infected with HIV. PLoS Pathog. 9, e1003328 (2013).
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nature Rev. Mol. Cell Biol. 8, 275–283 (2007).
Davids, M. S. & Letai, A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J. Clin. Oncol. 30, 3127–3135 (2012).
Maiuri, M. C. et al. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J. 26, 2527–2539 (2007).
Bellot, G. et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 29, 2570–2581 (2009).
Malik, S. A. et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 30, 3918–3929 (2011).
Andreu-Fernandez, V. et al. BH3-mimetics- and cisplatin-induced cell death proceeds through different pathways depending on the availability of death-related cellular components. PloS ONE 8, e56881 (2013).
Pattingre, S. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 (2005). Reveals the mechanisms through which BCL-2 can repress autophagy in addition to its anti-apoptotic function.
Schwarten, M. et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy 5, 690–698 (2009).
Luo, S. et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol. Cell 47, 359–370 (2012).
Wei, Y., Pattingre, S., Sinha, S., Bassik, M. & Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 30, 678–688 (2008).
Zalckvar, E. et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10, 285–292 (2009).
Eisenberg-Lerner, A. & Kimchi, A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 19, 788–797 (2012). References 43 and 45 unravel the mechanisms through which DAPK stimulate autophagy.
Gandesiri, M. et al. DAPK plays an important role in panobinostat-induced autophagy and commits cells to apoptosis under autophagy deficient conditions. Apoptosis 17, 1300–1315 (2012).
Guenebeaud, C. et al. The dependence receptor UNC5H2/B triggers apoptosis via PP2A-mediated dephosphorylation of DAP kinase. Mol. Cell 40, 863–876 (2010).
Widau, R. C., Jin, Y., Dixon, S. A., Wadzinski, B. E. & Gallagher, P. J. Protein phosphatase 2A (PP2A) holoenzymes regulate death-associated protein kinase (DAPK) in ceramide-induced anoikis. J. Biol. Chem. 285, 13827–13838 (2010).
He, C. et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 (2012).
del Peso, L., Gonzalez-Garcia, M., Page, C., Herrera, R. & Nunez, G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278, 687–689 (1997).
Wang, R. C. et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012).
Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J. & Codogno, P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998 (2000).
Scott, R. C., Schuldiner, O. & Neufeld, T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 (2004).
Iaccarino, I., Hancock, D., Evan, G. & Downward, J. c-Myc induces cytochrome c release in Rat1 fibroblasts by increasing outer mitochondrial membrane permeability in a Bid-dependent manner. Cell Death Differ. 10, 599–608 (2003).
Hart, L. S. et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J. Clin. Invest. 122, 4621–4634 (2012).
Guo, J. Y. et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25, 460–470 (2011).
Lock, R. et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 22, 165–178 (2011).
Wang, Y. et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc. Natl Acad. Sci. USA 109, 13325–13330 (2012). References 56 to 58 illustrate the importance of autophagy to survive stress occurring in cells expressing oncogenic RAS.
Schmukler, E. et al. Ras inhibition enhances autophagy, which partially protects cells from death. Oncotarget 4, 142–152 (2013).
Gomes, L. C., Di Benedetto, G. & Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature Cell Biol. 13, 589–598 (2011).
Kim, I. & Lemasters, J. J. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid. Redox Signal. 14, 1919–1928 (2011).
Grasso, D. et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1–USP9x–p62, prevents pancreatic cell death. J. Biol. Chem. 286, 8308–8324 (2011).
Fortunato, F. et al. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis. Gastroenterology 137, 350–360, (2009).
Shaid, S., Brandts, C. H., Serve, H. & Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 20, 21–30 (2013).
Hou, W., Han, J., Lu, C., Goldstein, L. A. & Rabinowich, H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 6, 891–900 (2010).
Amir, M. et al. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 20, 878–887 (2013).
Sandilands, E., Serrels, B., Wilkinson, S. & Frame, M. C. Src-dependent autophagic degradation of Ret in FAK-signalling-defective cancer cells. EMBO Rep. 13, 733–740 (2012).
Mathew, R. et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 (2009). Details the molecular and cellular mechanisms through which autophagy inhibition triggers carcinogenesis in the liver.
Oral, O. et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 17, 810–820 (2012).
Luo, S. & Rubinsztein, D. C. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 17, 268–277 (2010).
Wirawan, E. et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 1, e18 (2010).
Pagliarini, V. et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 19, 1495–1504 (2012).
Fujita, N. et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651–4659 (2008).
Betin, V. M. & Lane, J. D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 122, 2554–2566 (2009).
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biol. 8, 1124–1132 (2006). Illustrates how limited proteolysis of an autophagy-inducing gene product generates a protein with a new pro-apoptotic function.
Lamy, L. et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 23, 435–449 (2013).
Kroemer, G. & Levine, B. Autophagic cell death: the story of a misnomer. Nature Rev. Mol. Cell Biol. 9, 1004–1010 (2008).
Shen, S., Kepp, O. & Kroemer, G. The end of autophagic cell death? Autophagy 8, 1–3 (2012).
Galluzzi, L. et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 19, 107–120 (2012).
Shravage, B. V., Hill, J. H., Powers, C. M., Wu, L. & Baehrecke, E. H. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 140, 1321–1329 (2013).
Manjithaya, R. & Subramani, S. Autophagy: a broad role in unconventional protein secretion? Trends Cell Biol. 21, 67–73 (2011).
Young, M. M. et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 287, 12455–12468 (2012).
Nezis, I. P. et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190, 523–531 (2010).
Yu, L. et al. Autophagic programmed cell death by selective catalase degradation. Proc. Natl Acad. Sci. USA 103, 4952–4957 (2006).
Saftig, P., Beertsen, W. & Eskelinen, E. L. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy 4, 510–512 (2008).
Ma, X. et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation 125, 3170–3181 (2012).
Rubinstein, A. D., Eisenstein, M., Ber, Y., Bialik, S. & Kimchi, A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 44, 698–709 (2011).
Kessel, D. H., Price, M. & Reiners, J. J. Jr. ATG7 deficiency suppresses apoptosis and cell death induced by lysosomal photodamage. Autophagy 8, 1333–1341 (2012).
Boya, P. et al. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J. Exp. Med. 197, 1323–1334 (2003).
Deretic, V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104 (2011).
Tang, D., Kang, R., Coyne, C. B., Zeh, H. J. & Lotze, M. T. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 249, 158–175 (2012).
Zitvogel, L., Kepp, O. & Kroemer, G. Decoding cell death signals in inflammation and immunity. Cell 140, 798–804 (2010).
Huang, C. et al. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS ONE 6, e20975 (2011).
Perez-Pinzon, M. A., Stetler, R. A. & Fiskum, G. Novel mitochondrial targets for neuroprotection. J. Cereb. Blood Flow Metab. 32, 1362–1376 (2012).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007). A seminal paper demonstrating the impact of pre-mortem autophagy on the clearance of apoptotic cells.
Chekeni, F. B. et al. Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010).
Michaud, M. et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577 (2011). Evidence that inhibition of autophagy in tumour cells may subvert anticancer immunosurveillance and hence reduce the efficacy of chemotherapies.
Sukkurwala, A. Q. et al. Immunogenic calreticulin exposure occurs through a phylogenetically conserved stress pathway involving the chemokine CXCL8. Cell Death Differ. Jan 2013 (doi: http://dx.doi.org/10.1038/cdd.2013.73).
Bao, J. X. et al. Lysosome–membrane fusion mediated superoxide production in hyperglycaemia-induced endothelial dysfunction. PLoS ONE 7, e30387 (2012).
Fader, C. M., Aguilera, M. O. & Colombo, M. I. ATP is released from autophagic vesicles to the extracellular space in a VAMP7-dependent manner. Autophagy 8, 1741–1756 (2012).
Uhl, M. et al. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 16, 991–1005 (2009).
Ko, A. et al. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signaling. Cell Death Differ Sep 2013 (doi: http://dx.doi.org/10.1038/cdd.2013.124).
Ma, Y. et al. Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity 38, 729–741 (2013).
Calabrese, E. J., Iavicoli, I. & Calabrese, V. Hormesis: its impact on medicine and health. Hum. Exp. Toxicol. 32, 120–152 (2013).
Hetz, C. et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23, 2294–2306 (2009).
White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nature Rev. Cancer 12, 401–410 (2012).
Rello-Varona, S. et al. Autophagic removal of micronuclei. Cell Cycle 11, 170–176 (2012).
Kuo, T. C. et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nature Cell Biol. 13, 1214–1223 (2011).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Rubinsztein, D. C., Marino, G. & Kroemer, G. Autophagy and aging. Cell 146, 682–695 (2011).
Hamasaki, M. et al. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393 (2013).
Adams, J. M. & Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 (2007).
Kroemer, G., Galluzzi, L. & Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007). An exhaustive review on pro-survival and lethal signalling pathways converging on mitochondria, as well as on the mechanisms that decide whether mitochondrial membranes undergo deadly permeabilization.
Galonek, H. L. & Hardwick, J. M. Upgrading the BCL-2 network. Nature Cell Biol. 8, 1317–1319 (2006).
Kroemer, G. & Martin, S. J. Caspase-independent cell death. Nature Med. 11, 725–730 (2005).
Boya, P. & Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 (2008).
Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. & Kroemer, G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nature Rev. Mol. Cell Biol. 11, 700–714 (2010).
Vitale, I., Galluzzi, L., Castedo, M. & Kroemer, G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nature Rev. Mol. Cell Biol. 12, 385–392 (2011).
Samara, C., Syntichaki, P. & Tavernarakis, N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 15, 105–112 (2008). An important paper describing the contribution of autophagy to pathological loss of neurons in nematodes.
Erdelyi, P. et al. Shared developmental roles and transcriptional control of autophagy and apoptosis in Caenorhabditis elegans. J. Cell Sci. 124, 1510–1518 (2011).
Tracy, K. & Baehrecke, E. H. The role of autophagy in Drosophila metamorphosis. Curr. Top. Dev. Biol. 103, 101–125 (2013).
Berry, D. L. & Baehrecke, E. H. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 (2007). First genetic evidence that autophagic cell death occurrs during the development of Drosophila.
Denton, D. et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 19, 1741–1746 (2009).
Chang, T.-K. et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nature Cell Biol. 15, 1067–1078 (2013).
Gozuacik, D. et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 15, 1875–1886 (2008).
Byun, J. Y. et al. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis 30, 1880–1888 (2009).
Elgendy, M., Sheridan, C., Brumatti, G. & Martin, S. J. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 42, 23–35 (2011).
Koike, M. et al. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am. J. Pathol. 172, 454–469 (2008).
Zhu, H. et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 117, 1782–1793 (2007). An example of how autophagy may mediate deleterious effects in a model of pressure overload affecting the heart muscle.
Sun, Y. et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Science Signal. 5, ra16 (2012).
Sentelle, R. D. et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nature Chem. Biol. 8, 831–838 (2012).
Acknowledgements
The authors are supported by the Ligue contre le Cancer (équipe labellisée), Agence National de la Recherche, AXA Chair for Longevity Research, Association pour la Recherche sur le Cancer, Cancéropôle Ile-de-France, Institut National du Cancer (INCa), Fondation Bettencourt-Schueller, Fondation de France, Fondation pour la Recherche Médicale, the European Commission (ArtForce), the European Research Council, the LabEx Immuno-Oncology, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (Socrate), Cancer Research and Personalized Medicine (Carpem), the Paris Alliance of Cancer Research Institutes, the National Institutes of Health and the Ellison Medical Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Contribution of individual components of the Beclin 1 interactome to autophagy and apoptosis. (PDF 121 kb)
Glossary
- Autophagosomes
-
Vesicles characterized by two membranes that contain autophagic cargo.
- Autolysosomes
-
Fusion proteins of autophagosomes and lysosomes, also called autophagolysosomes. Autolysosomes only have one membrane.
- Programmed cell death
-
(PCD). Cell death that occurs during development or adult tissue homeostasis for the removal of superfluous cells.
- Mitochondrial outer membrane permeabilization
-
(MOMP). An apoptosis-associated process that results in the release of apoptosis-inducing proteins (such as cytochrome c, AIF (apoptosis-inducing factor), SMAC (second mitochondria-derived activator of caspase) and others) that are normally retained in the mitochondrial intermembrane space, through the outer membrane into the cytosol.
- Caspases
-
A family of Cys proteases that cleave after Asp residues. Initiator caspases are typically activated in response to a particular triggering event (for example, caspase 8 upon death receptor ligation, caspase 9 upon apoptosome activation and caspase 2 upon DNA damage), whereas effector caspases (mainly caspase 3, caspase 6 and caspase 7) are particularly important for the ordered dismantling of vital cellular structures.
- p53
-
A central tumour suppressor protein that has multiple functions, both in the cytoplasm and in the nucleus. As a transcription factor, p53 transactivates genes that induce cell cycle arrest, cellular senescence, autophagy and apoptosis.
- Lysosomal membrane permeabilization
-
(LMP). A disruption of lysosomal membrane function that leads to the translocation of lysosomal hydrolases, including cathepsins, from the lysosomal lumen to the rest of the cell. LMP can be induced by endogenous signal transducers (such as reactive oxygen species and sphingosine), as well as by lysosomotropic drugs.
- BCL-2 family
-
Each member of this protein family contains at least one BCL-2 homology (BH) region. The family is divided into anti-apoptotic multidomain proteins (such as BCL-2, BCL-XL (BCL extra large) and MCL1 (myeloid cell leukaemia sequence 1)), which contain four BH domains (BH1, BH2, BH3 and BH4), pro-apoptotic multidomain proteins (for example, BAX (BCL-2- associated X protein) and BAK (BCL-2 antagonist or killer)), which contain BH1, BH2 and BH3, and the pro-apoptotic BH3-only protein family.
- Unfolded protein response
-
(UPR). The UPR is activated in response to stress in the endoplasmic reticulum (ER). It enables cells to adapt to ER stress by reducing the quantity of misfolded proteins in this organelle.
- Senescence
-
An irreversible G1 cell cycle arrest that is accompanied by morphological changes (flattening of the cells), metabolic alterations and transcriptional reprogramming that leads to the expression of cell cycle blockers, such as p16 and p21, and senescence-associated ß-galactosidase.
- Cytochrome c
-
A haem protein exclusively present in the mitochondrial intermembrane space. During the initiation of apoptosis, cytochrome c is released from mitochondria and triggers the assembly of the apoptosome, which is a caspase activation complex.
- Reactive oxygen species
-
(ROS). Classic oxygen radicals and peroxides that are formed within cells.
- Death receptors
-
Cell surface receptors that activate the extrinsic pathway of apoptosis upon ligand-induced trimerization. The family of death receptors includes CD95 (which binds CD95 ligand), tumour necrosis factor receptor 1 (TNFR1) and two receptors for TNF-related apoptosis-inducing ligand (TRAILR1 and TRAILR2).
- Sphingosine kinase
-
A conserved lipid kinase that catalyses the formation of the bioactive sphingolipid metabolite sphingosine 1- phosphate. This acts as a second messenger, with important roles in numerous physiological processes, including cell growth, motility and survival.
- Danger-associated molecular patterns
-
(DAMPs). These molecules are released by or exposed on the surface of stressed and dying cells. They initiate and perpetuate sterile inflammatory responses.
- Pattern recognition receptors
-
(PRRs). A series of intracellular or surface receptors that sense danger-associated molecular patterns (DAMPs) or foreign structures from infectious pathogens, triggering inflammatory and immune responses.
- Heterophagy
-
Phagocytosis of a cell by another cell. Heterophagy has a major role in the efficient removal of apoptotic cells. Efficient heterophagy is indispensable for avoiding inflammatory responses triggered by apoptotic material.
- Amyotrophic lateral sclerosis
-
(ALS). A progressive and debilitating neurodegenerative disease that is characterized by progressive muscle atrophy and other degenerative manifestations. The disease pathophysiology is complex and not yet fully understood, but it is proposed to involve the accumulation of misfolded proteins.
Rights and permissions
About this article
Cite this article
Mariño, G., Niso-Santano, M., Baehrecke, E. et al. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15, 81–94 (2014). https://doi.org/10.1038/nrm3735
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3735
This article is cited by
-
Icariin exerts anti-tumor activity by inducing autophagy via AMPK/mTOR/ULK1 pathway in triple-negative breast cancer
Cancer Cell International (2024)
-
Modulation of autophagy and apoptosis can contribute to the anticancer effect of Abemaciclib/Celecoxib combination in colon cancer cells
Medical Oncology (2024)
-
Transcription factor EB-mediated autophagy affects cell migration and inhibits apoptosis to promote endometriosis
Apoptosis (2024)
-
Protective effect of hygrolansamycin C against corticosterone-induced toxicity and oxidative stress-mediated via autophagy and the MAPK signaling pathway
Pharmacological Reports (2024)
-
Autophagic degradation of MVBs in LSECs promotes Aldosterone induced-HSCs activation
Hepatology International (2024)