Abstract
A significant variation in susceptibility to paclitaxel-mediated killing was observed among a panel of short-term cultured non-small-cell lung cancer (NSCLC) cell lines. Susceptibility to killing by paclitaxel correlated with expression of the BH3-only protein, Bim, but not with other members of Bcl-2 family. NSCLC cell lines with the highest level of Bim expression are most susceptible to apoptosis induction after paclitaxel treatment. Forced expression of Bim increased paclitaxel-mediated killing of cells expressing an undetectable level of Bim. Conversely, knock down of Bim, but not Bcl-2 expression, decreased the susceptibility of tumor cells to paclitaxel-mediated killing. Similar observations were made using a panel of breast and prostate cancer cell lines. Paclitaxel impairs microtubule function, causes G2/M cell cycle blockade, mitochondria damage, and p53-independent apoptosis. These results established Bim as a critical molecular link between the microtubule poison, paclitaxel, and apoptosis.
Similar content being viewed by others
Introduction
The taxanes, paclitaxel and docetaxel, are among the most effective chemotherapeutic agents for patients with NSCLC, prostate and breast cancer.1 Taxanes induce stabilization of microtubule formation, blockade of G2/M cell cycle transition, leading to subsequent cell death. The cell death pathways in cancer cells targeted by taxanes are not well-defined. In contrast to DNA-damaging agents, which induce p53-dependent apoptosis, antimicrotubule agents, including paclitaxel, induce apoptosis regardless of the status of p53 and loss of p53 appears to sensitize cells to cell death stimuli.2, 3, 4, 5, 6 Two major cell death pathways have been described, one mediated by death receptors and the other mediated by mitochondria. Both pathways involve a signal cascade of caspases. For example, the initiator caspases, caspase-8, or -9, are directly activated by death receptors or cytochrome c released from mitochondria, and subsequently transmit the signal to downstream effector caspases, such as caspase-3, -6, or -7.7 It is well established that the intrinsic apoptotic pathway mediated by mitochondria is critical for cell death induced by various forms of cell stress including the perturbation of microtubule dynamics inflicted by paclitaxel.8, 9, 10, 11, 12, 13 This intrinsic cell death pathway is largely regulated by both pro- and antiapoptotic members of the Bcl-2 family.14 BH3-only proapoptotic proteins are localized in different cellular compartments and situated to sense different sources of stress, whereas antiapoptotic proteins such as Bcl-2 and Bcl-xL are localized on the outer membrane of mitochondria, guiding the integrity of mitochondrial outer membrane. Previous studies demonstrated that paclitaxel induced phosphorylation of Bcl-22 was critical for cell death induced by paclitaxel.15, 16 However, more recent studies showed that Bcl-2 phosphorylation occurs normally during the G2/M phase of the cell cycle17 and raised doubt about whether Bcl-2 phosphorylation was the cause of cell death or merely a bystander event. In addition, there was poor correlation between paclitaxel resistance and expression of Bcl-2 and Bcl-xL in lung cancer cell lines; overexpression of Bcl-2 failed to protect NSCLC cell lines from paclitaxel-induced apoptosis.18 Recently, the BH3-only proapoptotic protein Bim was shown to localize to microtubules via binding to the light chain of the dynein motor complex19 and to be critical for paclitaxel-mediated cell death of lymphocytes,20 suggesting that Bim may be an important mediator of cell death induced by antimicrotubule agents in solid tumors.
Results
Expression of Bcl-2 family members and susceptibility to paclitaxel killing
We recently established a panel of short-term cultured cell lines from chemotherapy naïve NSCLC patients (LuCSF1, LT-46, LT-30, LT-36, CRI-702, and CRI-619). To determine their susceptibility to paclitaxel, cells were treated with 10 to 80 nM paclitaxel for 24–72 h and viability was analyzed by propidium iodide (PI) staining and flow cytometry. At 48 h, CRI-702 and CRI-619 were the most sensitive to paclitaxel-induced killing compared to four other cell lines (Figure 1a). To determine whether differential susceptibility of these cell lines to paclitaxel was related to differential expression of Bcl-2 family members, expression levels of Bim, Bmf, Bax, Bak, Bcl-2, and Bcl-xL were measured by Western blot analysis. Their protein levels varied greatly among these cell lines (Figure 1b). Interestingly, the two paclitaxel-sensitive cell lines (CRI-702 and CRI-619) expressed high levels of Bim (predominantly the BimEL isoform), while four resistant cell lines (LuCSF1, LT-30, LT-46 and LT-36) expressed a significant lower level of Bim (Figure 1b). In agreement with published reports, expression of Bmf, Bax, Bak, Bcl-2, or Bcl-xl did not correlate with the susceptibility to paclitaxel (Figure 1b). For example, the sensitive lines, CRI-619 and CRI-702, expressed the highest levels of Bcl-2 proteins. After 40 nM paclitaxel treatment, expression of BimEL isoform in paclitaxel-sensitive cell line CRI-619 decreased, while expression of BimL isoform increased. Most importantly, the most apoptotic BimS isoform was expressed only after paclitaxel treatment (Figure 1c). In contrast, a slight induction of BimEL isoform was found in paclitaxel resistant cell line LuCSF1. The increased expression of BimL isoform and the de novo synthesis of BimS isoform were found only in detached cell population. Killing of NSCLC cell lines by paclitaxel appeared to be independent of protein neosynthesis, since actinomycin D treatment only slightly enhanced paclitaxel-mediated killing of sensitive cell lines and did not affect resistant cell lines (data not shown). To extend the connection between the level of Bim expression and paclitaxel-mediated tumor killing to other cancer types, the experiments were repeated with four breast (SK-BR-3, BrCa101, BccMcGee, and MDA) and two prostate (DU145 and PC3) cancer cell lines. Again, tumor cell lines that expressed higher level of Bim exhibited greater level of sensitivity to paclitaxel (Figure 2). These results suggest that, as in lymphocytes, paclitaxel-mediated killing of human carcinoma cells may also depend on Bim expression.
Paclitaxel-mediated killing and expression of Bcl-2 family members in NSCLC cell lines. (a) Six NSCLC cell lines were incubated with different concentration of paclitaxel. Cell death was determined by PI staining and FACS analysis after 48 h treatment. Both attached and detached cells were collected for the FACS analysis. (b) Expression of Bcl-2 family members in NSCLC cell lines by Western blot analysis. Cell lysates were prepared from all six cell lines. The protein concentration of cell lysate was determined by Bradford method. An equal amount of total protein (10 μg per lane) was loaded into each lane. (c) Expression of Bim isoforms before and after 40 nM paclitaxel treatment in CRI-619 and LuCSF1 cell lines. Cell lysate was prepared from cells without paclitaxel treatment and 24 and 48 h after paclitaxel treatment. For cells treated with paclitaxel, lysates were prepared from attached and detached cells or cells combined of both population. The protein concentration of cell lysate was determined by Bradford method. An equal amount of total protein (10 μg per lane) was loaded into each lane. Also, β-actin was detected as a control
Paclitaxel-mediated killing and expression of Bcl-2 family members in breast and prostate cell lines. (a) Four breast and two prostate carcinoma cell lines were treated with indicated concentration of paclitaxel for 48 h, both attached and detached cells were then collected and stained with PI and cell viability was determined by flow cytometry analysis. (b) Expression of Bcl-2 family members in prostate and breast cancer cell lines by Western blot analysis. The protein concentration of cell lysate was determined by Bradford method. An equal amount of total protein (10 μg per lane) was loaded into each lane
Paclitaxel induced apoptosis, mitochondrial damage, and caspase activation
To determine whether exposure to paclitaxel leads to caspase activation and apoptosis, CRI-702 cells were incubated with 40 nM paclitaxel for 24 h. Cells treated with paclitaxel die by apoptosis, which is manifested morphologically by membrane bubbling, nuclear condensation, and fragmentation (Figure 3a). Caspase activation was assessed by using fluorogenic substrates specific for caspase-2, -3, -8, and -9; paclitaxel sensitive (CRI-702) but not resistant cells (LuCSF1) exhibited a significant increase of caspase-2, -3, -8, and -9 activation (Figure 3b). The activation of caspase-3 was confirmed by Western blot analysis of the cleavage of procaspase-3, which predominantly occurred in detached cells (Figure 3c). Furthermore, paclitaxel treatment caused a dramatic loss of mitochondrial membrane potential in CRI-619 as measured by the fluorescent probe JC-1 (Figure 3d). Thus, paclitaxel could induce apoptotic cell death in sensitive NSCLC cell lines.
Paclitaxel-induced apoptosis and caspase activation in the NSCLC cell line CRI-702. (a) CRI-702 cells exhibited apoptotic changes after exposure to paclitaxel. Cells were incubated with 40 nM paclitaxel for 24 h and examined directly under the microscope in bright field, or after staining with the cell-permeable nuclear stain HS-33342, or calcein-AM and EthD-1. (b) Activation of caspases was observed in CRI-702 cells but not LuCSF1 cells. After 48 h exposure to paclitaxel, cell lysate was prepared and caspase activation was determined using caspase-specific fluorescent AFC substrates. Induction fold was calculated as AFC fluorescence with paclitaxel treatment minus background fluorescence divided by AFC fluorescence without paclitaxel treatment minus background. (c) Paclitaxel induced cleavage of procaspase-3 in CRI-702 cell line. CRI-702 cells were treated with 40 nM paclitaxel and cell lysate was prepared from detached cells, attached cells, or the total cell population after 24 or 48 h of exposure. The cleavage of procapase-3 was determined by Western blot analysis with antibody against caspase-3. Each lane was loaded with 10 μg of protein lysate. (d) Loss of mitochondrial membrane potential after exposure to paclitaxel. CRI-619 cells were treated with 40 nM paclitaxel for 24 h and stained with JC-1 and analyzed by flow cytometry. The numbers represent the percentage of cells that lost mitochondrial potential as indicated disappearance of JC-1 aggregates and appearance of JC-1 monomers
Caspase inhibitors reduced paclitaxel-induced nuclear condensation
Paclitaxel has been shown to induce mitochondrial damage and apoptosis in both caspase-dependent and -independent manner in various cell lines.18, 21, 22, 23, 24To determine whether apoptosis of our NSCLC cell lines is caspase-dependent, CRI-619 and CRI-702 cells were incubated with cell-permeable inhibitors to pancaspases, caspase-3, -8, and -9, and then treated with 40 nM paclitaxel for 24 h. As measured by HS-33342 nuclear staining, a low but significant blockage of nuclear fragmentation was observed in CRI-702 cells (Figure 4a); inhibitors to pancaspases and casapse-3 and -8 but not caspase-9 significantly decreased paclitaxel-induced apoptosis. Apoptosis of CRI-619 was greatly diminished in the presence of inhibitors to pancaspases or caspase-3 but not caspase-8 or -9 (Figure 4b). In all experiments, control inhibitor Z-FA-FMK did not change paclitaxel susceptibility of either cell line. These results suggest that caspases are important for paclitaxel-induced apoptosis and the requirement for which particular caspase may depend on the cell line tested. We observed approximately four-fold induction of caspase-3-like activity using DEVD substrate and a marked effect on survival after pretreatment with DEVD inhibitor; however, procaspase-3 was processed incompletely (Figure 3c). This indicated that an additional executor caspase or caspases, such as caspase-7, might also involve in paclitaxel-mediated apoptosis.
Effect of inhibition of caspases on paclitaxel-induced apoptosis. Paclitaxel-sensitive CRI-702 (a) and CRI-619 (b) cells were first incubated with 50 μM control inhibitor (Z-FA-FMK), and pan-caspase (Z-VAD-FMK), caspase-3 (Z-DEVD-FMK), -8 (Z-IETD-FMK), and -9 (Z-LEHD-FMK) inhibitors for 40 min before the addition of 40 nM paclitaxel. After 24 h, Hoechst nuclear staining was used to enumerate apoptotic nuclei. Each image contained 50–100 cells, and the percentage of apoptotic cells was calculated by dividing the number of cells with bright and small nuclei by the total number of cells. (a) Apoptosis of CRI-702 cells were reduced by pancaspase inhibitors or caspase-3 and -8 selective inhibitors, but not caspase-9 inhibitor. (b) Pancaspase and caspase-3 inhibitors, but not caspase-8 and -9 inhibitors, reduced paclitaxel-induced apoptosis of CRI-619 cells
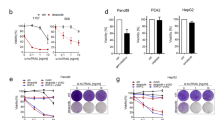
Forced expression of Bim increased paclitaxel susceptibility of LuCSF1 cell line
The maximum level of paclitaxel-mediated killing of LuCSF1 cells is relatively low. If it is due to the lack of Bim expression, forced expression of Bim by a retroviral vector would lead to an increased susceptibility to paclitaxel. To test this hypothesis, LuCSF1 cells were transduced with retroviral vectors that encode BimL and GFP proteins or GFP alone.25 After sorting, Bim expression was confirmed by immunoprecipitation and Western blot analysis (Figure 5a). Consistent with our hypothesis, the expression of Bim significantly increased the susceptibility of tumor cells to paclitaxel (Figure 5b). The increased killing was accompanied by increased levels of caspase-3-like activity induced by paclitaxel treatment (Figure 5c). Thus, one potential mechanism for paclitaxel resistance is the lack of Bim expression in cell lines such as LuCSF1.
Transduction of LuCSF1 cells with Bim retroviral vector increased susceptibility to paclitaxel-mediated killing. (a) LuCSF1 cells were transduced with pMIG-Bim or pMIG retroviruses and selected for GFP expression by cell sorting. Expression of Bim and Bak was determined by immunoprecipitation and Western blot analysis (Bim) or Western blot analysis (Bak). (b) Expression of exogenous Bim-sensitized LuCSF1 cells to paclitaxel-mediated killing. Nontransduced GFP or Bim-transduced LuCSF1 cells were treated with paclitaxel for 48 h at different concentrations indicated. Cell viability was measured by PI staining and FACS analysis. (c) Paclitaxel induced a higher level of caspase-3-like activity in cell lines expressing exogenous Bim. Nontransduced or transduced LuCSF1 cell lines were treated with 10 nM paclitaxel for 48 h and the caspase-3-like activity was determined with fluorescent substrate Ac-DEVD-AFC
Transient knockdown Bim expression by small interfering RNA (siRNA) decreased paclitaxel-mediated killing of breast, prostate, and NSCLC cell lines
To confirm that Bim is required for paclitaxel-mediated killing of tumor cells, the sensitivity to paclitaxel of four tumor cell lines (MDA, SK-BR-3, CRI-702, and PC3) was determined after Bim expression was transiently knocked down by Bim-specific siRNA using lipofection. Bim, but not control GFP, siRNA was able to reduce Bim expression in all four cell lines; Bak expression was not affected by either siRNA (Figure 6a). A significant level of decrease in killing was observed following Bim siRNA transfection compared to GFP siRNA in all cell lines tested (Figure 6b). Transfection with siRNA alone did not significantly change the basal level of cell death in all cell lines tested (Figure 6c).
Transient Bim knock down with specific siRNA reduced susceptibility to paclitaxel-mediated killing. (a) MDA, SK-Br-3, CRI-702, and PC3 cells were transfected with Bim or GFP siRNA (0.25 μg/ml siRNA for CRI-702, 0.5 μg/ml siRNA for MDA, SK-Br-3, and PC3) using Lipofectamine 2000 reagent. At 24 h post-transfection, Bim and Bak expression were determined by Western blot analysis. (b) Transient knockdown Bim expression decreased paclitaxel-mediated killing of NSCLC, breast, and prostate cancer cells. At 24 h after siRNA lipofection, cells were treated with 40 nM paclitaxel for 48 h. The cell viability was determined by PI staining and FACS analysis. Each experiment was repeated three times. The data represent typical results from three independent experiments. (c) Transfection with GFP or Bim siRNA did not significantly change the basal level of cell death in CRI-702, MDA, SK-Br-3, and PC3 cell lines. Spontaneous cell death was measured in nontransduced and transduced cells by PI staining and FACS analysis. Each experiment was repeated three times. The data represent typical results from three independent experiments
Knockdown Bcl-2 expression by siRNA did not affect paclitaxel-mediated killing of breast cancer and NSCLC cell lines
Two most sensitive cell lines, CRI-702 and CRI-619, expressed high level of Bim as well as Bcl-2 (Figure 1b). It has been reported previously that phosphorylation and inactivation of Bcl-2 played a critical role in paclitaxel-mediated killing.15, 16 As phosphorylation and inactivation of Bcl-2 occurs during the G2/M phase of normal cell cycle,17 it remains controversial whether Bcl-2 is required for paclitaxel-mediated killing. To determine whether paclitaxel-mediated killing of tumor cells requires expression of Bcl-2, knockdown experiments were repeated with Bcl-2 siRNA by lipofection. Bcl-2, but not control GFP, siRNA was able to reduce Bcl-2 expression in all three cell lines; Bak expression was not affected by either siRNA (Figure 7a). In contrast to Bim siRNA, no significant difference of killing was observed following transfection with either Bcl-2 or GFP siRNA (Figure 7b). Once again, transfection with siRNA alone did not significantly change the basal level of cell death in all cell lines (Figure 7c).
Bcl-2 knockdown with specific siRNA did not affect the susceptibility to paclitaxel-mediated killing. (a) Bcl-2, but not Bak, was specifically knocked down in breast and NSCLC cell lines by the Bcl-2 siRNA. Bcl-2 and GFP siRNA were transfected into breast cancer (MDA) and NSCLC (CRI-619 and CRI-702) cell lines by lipofection. The expression of Bcl-2 and Bak was determined by Western blot analysis. (b) No significant difference of sensitivity to paclitaxel was observed in cell lines following Bcl-2 and GFP knock down. At 24 h after siRNA lipofection, cells were treated with 40 nM paclitaxel for 36 h. The cell viability was determined by PI staining and FACS analysis. Each experiment was repeated twice. The data represent typical results from two independent experiments. (c) Transfection with GFP or Bim siRNA did not significantly change the basal level of cell death in CRI-619, CRI-702, and MDA cell lines. Spontaneous cell death was measured in nontransduced and transduced cells by PI staining and FACS analysis. Each experiment was repeated twice. The data represent typical results from two independent experiments
Stable knockdown Bim expression decreased sensitivity to paclitaxel-mediated killing in CRI-619 cell line
In previous experiments, we used the transient lipofection method to deliver siRNA into tumor cells. We observed that the control GFP siRNA sometimes could reduce the killing mediated by paclitaxel, albeit at a lower level compared to Bim siRNA. This could be due to the activation of PKR or other signaling pathways that often associated with transient transfection. To overcome this problem, we generated stable knockdown cell lines using retroviral vector system. After selection with puromycin, we established two cell lines derived from CRI-619, CRI-619 Bim siRNA and CRI-619 luc siRNA. CRI-619 tumor cells expressing Bim siRNA, but not control luciferase siRNA, decreased BimEL and BimL expression, while Bak expression was not affected by either siRNA (Figure 8a). Consistent with results obtained from transient knockdown experiments, a significant decrease in killing levels was observed in CIR-619 Bim siRNA compared to the control CIR-619 luc siRNA or parental CRI-619 cells (Figure 8b). The levels of killing were also determined with HS-33342 staining assay and similar results were obtained (data not shown).
CRI-619 expressing stable Bim siRNA increased resistance to paclitaxel-mediated killing. (a) Decreased expression of Bim, but not Bak, in CRI-619 tumor cells that express stable Bim siRNA. The expression of Bim and Bak was determined by Western blot analysis. (b) Stable reduction of Bim expression significantly decreased the sensitivity to paclitaxel-mediated killing. CRI-619, CRI-619 luc siRNA and CRI-619 Bim siRNA were treated with paclitaxel at different concentration for 48 h. The cell viability was determined by PI staining and FACS analysis
Discussion
Paclitaxel, cell cycle arrest, and apoptosis
It is well established that antimicrotubule drugs such as paclitaxel induce G2/M arrest by activating spindle checkpoints.26, 27 Cell death induction by antimicrotubule drugs requires the spindle check point signals. Treated with a low concentration of paclitaxel, checkpoint-deficient cells become polyploid and survive, while checkpoint-proficient cells exit mitosis and enter G1 followed by apoptosis.24, 28 Recent studies strongly support this notion, since amplification of Aurora-A leads to paclitaxel resistance by over-riding the mitotic spindle checkpoint.29 It was also found that mitotic spindle checkpoint-deficient lung cancer cells were highly resistant to antimicrotubule agents and checkpoint-proficient lung cancer cells became highly resistant when mitotic arrest was abrogated by treatment with staurosporine.30 The link between the activation of spindle checkpoint and subsequent cell death is unknown.
Taxanes and new classes of microtubule-stabilizing drugs, discodermolide and epothilone B, have been shown to induce caspase and mitochondrial-independent apoptosis in NSCLC, ovarian, and breast cancer cells.18, 31 Bim has a critical role in lymphocyte apoptosis induced by both cytokine withdrawals20 and TCR stimulation,32 and is required for neuronal cell death after NGF deprivation.33 Paclitaxel-induced apoptosis of lymphoma cells appeared to be caspase-dependent and involved mitochondria amplification loop.34 Our results showed that Bim-dependent apoptosis induction in sensitive cell lines was at least partially caspase-dependent. The most likely explanation for the discrepancy between our results and other reports may be explained by differences in the cell lines used. Our study investigated apoptosis in two cell lines that were relatively sensitive to paclitaxel, while paclitaxel induced a less efficient apoptosis in the H460 cells used in their study. Resistance to chemotherapy was often associated with dysfunction of mitochondrial death machinery and caspase inhibition.35, 36, 37, 38 In these cell lines, cell death may rely on other less-defined pathways when the major cell death mechanisms are either missing or blocked. It has been reported that apoptosis induction in caspase-inefficient NSCLC cells may involve an AIF-mediated and caspase-independent mechanism.38 Caspase-independent necrosis-like cell death can occur via serine proteases39 or via the hypoxia-inducible BH3-only proapoptotic protein Bnip3.40, 41 Unlike other BH3-only proteins, Bnip3-induced apoptosis required neither Apaf-1/cytochrome c nor caspases.40, 41 However, the participation of such pathways in paclitaxel induced-cell death has not been explored. Nevertheless, the ability of IAP antagonist Smac/Diablo to sensitize cells to paclitaxel-induced apoptosis strongly supports the notion that caspase activation is important for the robust cell death in sensitive cell lines and resistant cell lines after neutralization of caspase inhibitors.22, 34, 42, 43, 44 In addition, it is worth noting that activation of p34cdc2 cyclin kinase by paclitaxel leads to increased expression of survivin, a protein that regulates mitosis and inhibits apoptosis; blocking survivin expression by inhibiting p34cdc2 kinase resulted in massive mitochondrial-mediated apoptosis.45
Regulation of Bim
Bim is expressed by cells of hematopoietic, neuronal, and epithelial lineage.46 Three major isoforms of Bim (BimEL, BimL, and BimS) exist due to alternative splicing.47 Regulation of Bim has been reported at both transcriptional and post-transcriptional levels. In neuronal and other type cells, Bim expression was shown to be controlled by the FOXO3 transcriptional factor, which involved Akt/PKB.48, 49 Currently, it is not known what regulates Bim expression and function in prostate and NSCLC cell lines. It will be interesting to examine whether expression of Bim in prostate and NSCLC cell lines is also regulated by FOXO3 or other related members as recently shown for breast cancer cell lines.50 Our current study only examined the constitutive expression of Bim by different cell lines; the potential upregulation of Bim by paclitaxel treatment may also play a critical role in apoptosis induction. In fact, we found that paclitaxel could induce a high level of de novo expression of BimS isoform in sensitive CRI-619 and upregulation in CRI-702 cells. BimS isoform is the most potent inducer of apoptosis due to the lack of the dynein light-chain binding domain (present in both BimEL and BimL isoforms) and phosphorylation sites for Erk MAPKs (present only in BimEL isoform). Regulation of alternative splicing of Bim by paclitaxel has not been studied and will be one interesting area for future investigation.
The proapoptotic activity of Bim was also found to be regulated by the MAPKs at post-transcriptional levels. Two different but inter-related mechanisms have been proposed for the post-transcriptional regulation of Bim: sequestration to microtubules via binding to the light chain of the motor complex19 and JNK-dependent phosphorylation.51, 52 Lei et al. showed that BimL and Bmf were normally sequestered by binding to dynein and myosin V motor complexes and phosphorylation by JNK caused their release and mediated apoptosis induction by UV light.52 The phosphorylation site (Thr56) of BimL is within the binding site of dynein light chain, LC8. It is clear that Bim phosphorylation by JNK played an important role in neuronal cell death, although phosphorylation of BimEL was found to be at a different one, Ser65, which is outside the LC8 binding site.51 However, it is not clear whether release from the sequestration of Bim by the dynein complex played any significant role in neuronal cell death since induced BimEL was localized exclusively to mitochondria. Mostly importantly, it is still not known how phosphorylation of Bim might promote the proapoptotic activity of BimL or Bim EL.
Bim is regulated also at the post-translational level. While JNK appeared to regulate Bim activation, Erk members of MAPK family have been shown to modulate Bim protein levels via proteasome degradation pathways. It has been recently demonstrated that phosphorylation of BimEL by Erk1/2 on Ser65 selectively leads to its proteasome degradation.53, 54, 55 Recent studies have demonstrated that EGFR prevented anoikis by blocking induction of Bim expression in breast cancer cell lines via Erk-dependent pathways.56 As paclitaxel induces activation of MAPKs,57, 58, 59, 60 it will be important to determine whether the differential expression of Bim observed is caused by different levels of constitutive Erk1/2 phosphorylation associated with the cancer cells examined.61
Summary
Antimicrotubule drugs represent an important class of chemotherapeutics for various solid tumors. The underlying mechanisms by which these drugs induce mitochondria-mediated apoptosis are largely unknown. This report represents an important initial step in elucidation of such mechanisms. Future studies are needed to identify the potential molecular links between the activation of the mitotic spindle checkpoint and activation of Bim by JNK. In addition, elucidation of the mechanisms that regulate expression and proapoptotic activity of Bim in cancer cells will yield important information for the development of novel chemotherapeutic drugs and combination strategies.
Materials and Methods
Cell culture and paclitaxel killing
The human NSCLC cell lines (LuCSF1, LT-46, LT-30, LT-36, CRI-702, and CRI-619) and two breast cancer cell lines (BrCa-101 and BccMcGee) were short-term cultured cell lines derived from patients. Two breast cancer cell lines (MDA-MB-231 and SK-Br-3) and two prostate cancer cell lines (PC3 and Du145) were bought from ATCC (American Type Culture Collection). All tumor cells were cultured in RPMI-1640 media with 10% FBS and supplemented with nonessential amino acid and glutamine (BioWhittaker, MD, USA). Tumor cells were passaged two to three times per week by brief trypsinization and maintained in T-150 culture flasks. Paclitaxel (Sigma, St. Louis, MO, USA) was used at the indicated concentrations. For killing assays, cells were plated at a density of 0.5 × 106 cells in six-well plates overnight before replacing with media containing different concentrations of paclitaxel and incubated for 24, 48, or 72 h. Cell viability was determined by FACS analysis with PI staining (Sigma, MO, USA). The viability of tumor cells without exposure to paclitaxel was used as the control.
Western blot analysis
Cell lysates were prepared and quantified by the BCA method as described previously.62 Protein (10 μg/lane) was resolved by SDS-PAGE and transferred to PVDF (polyvinylidene difluoride) membranes according to the manufacturer's instructions (Invitrogen Life Technology Inc., Carlsbad, CA, USA). The membranes were blotted with antibodies specific for Bim (Oncogene, Boston, MA, USA), Bcl-xL (eBioscience, San Diego, CA, USA), Bak, Bcl-2, Bax (Upstate Biotechnology, Lake Placid, NY, USA), and β-actin (Sigma, MO, USA), and detected with anti-mouse IgG-HRP (Molecular Probes, Eugene, OR, USA) and ECL reagents as recommended by the manufacturer (Amersham Biosciences, Piscataway, NJ, USA). In some experiments, Bim was immunoprecipitated first before Western blot analysis to increase the sensitivity of detection.
Retroviral transduction
Tumor cells were transduced with the murine stem cell virus pMIG-BimL virus or with the pMIG control virus as described previously.62 Briefly, retroviral supernatants were prepared by transient transfection of 293T cells with pMIG control vector or pMIG-BimL vector with packaging vector pPack-GP and pPack-G encoding VSV-G envelope protein. Target cells were infected twice with retroviral supernatant in the presence of 8 μg/ml polybrene. Transduced cells were sorted using the MoFlow as GFP-positive cells and expanded in culture.
Bim and Bcl-2 knock down by siRNA
Bim, Bcl-2, and GFP siRNA were prepared as described (esiRNA).63 The forward primer of BimL with T7 sequence is TAA TAC GAC TCA CTA TAGG GAG ACA GGA TGG CAA AGC AAC CTT CTG. The reverse primer of BimL with T7 sequence is TAA TAC GAC TCA CTA TAGG GAG ACA GGT CAA TGC ATT CTC CAC AC. The forward primer of Bcl-2 with T7 sequence is TAA TAC GAC TCA CTA TAGG ATG GCG CAC GCT GGG AGA AC. The reverse primer of Bcl-2 with T7 sequence is TAA TAC GAC TCA CTA TAGG TCA CTT GTG GCC CAG ATA. Briefly, the full-length double-strand RNA was prepared by the in vitro transcription from BimL, Bcl-2, and GFP cDNA, and digested with RNase III to yield 15–23 bp small RNA. Cells (2 × 105 cells/well in six-well plate) were transfected twice with Bim, or Bcl-2 or GFP siRNA (0.25 μg/ml siRNA for CRI-702, 0.5 μg/ml siRNA for SKBr3, MDA, and PC3), using Lipofectamine 2000 reagent (Invitrogen Life Technology Inc., CA, USA) following the manufacturer's protocol. At 24 h post-transfection, Bim, Bcl-2, and Bak expression of transfected cells was determined by Western blot analysis and susceptibility to killing by paclitaxel was determined as above.
Establish stable CRI-619 cell line expressing stable Bim siRNA
To establish a permanent Bim knockdown cell line, we used BDTM knockout RNAi system (BD Biosciences, San Jose, CA, USA). The pSIREN-RetroQ vector is a retroviral shRNA expression vector that contains a puromycin resistance gene for the selection of stable transfectants. We constructed the Bim RNAi-Ready pSIREN vector according to the manufacturer's protocol. Briefly, annealed oligonucleotide (Bim RNAi sense: GAT CCG ACC GAG AAG GTA GAC AAT TGT TCA AGA GAC AAT TGT CTA CCT TCT CGG TCT TTT; Bim RNAi antisense: AAT TCG CTA GCA AAA AAG ACC GAG AAG GTA GAC AAT TGT CTC TTG AAC AAT TGT CTA CCT) was ligated into pSIREN vector. Retroviral supernatants were prepared by transient transfection of 293T cells with pSIREN Bim vector and pSIREN luciferase vector (provided by the manufacturer) with packaging vector pPack-GP and pPack-G. CRI-619 cells were infected twice with retroviral supernatant and stable cell lines were established by selection with 10 μg/ml puromycin (InvivoGen, San Diego, CA, USA). CRI-619, CRI-619 luc siRNA and CRI-619 Bim siRNA were treated with paclitaxel at different concentrations for 48 h. Cell viability was determined by FACS analysis with PI staining (Sigma, MO, USA).
Caspase activation and inhibition assays
The caspase substrates, Ac-VDVAD-AFC, Ac-DEVD-AFC, Ac-IETD-AFC, and Ac-LEHD-AFC, were used to measure caspase-2, -3, -8, and -9 activities, respectively, as recommended by the manufacturer (Pharmingen, CA, USA). Briefly, after incubation with paclitaxel, both the floating cells and adherent cells were collected and lysed in 150 μl of RIPA lysis buffer. For each reaction, 50 μl lysate and 5 μl reconstituted substrate were incubated for 1 h at 37°C in a 96-well plate. AFC fluorescence intensity was measured using a Wallac 1420 Victor2 with an excitation wavelength and emission set at 355 and 460 nM, respectively. Reconstituted substrate plus HEPES buffer alone was used to determine the background level of AFC fluorescence intensity. Induction fold was calculated as AFC fluorescence with paclitaxel treatment minus background fluorescence divided by AFC fluorescence without paclitaxel treatment minus background. For inhibition of caspases, cells were first incubated with 50 μM control inhibitor (Z-FA-FMK), and pan-caspase (Z-VAD-FMK), caspase-3 (Z-DEVD-FMK), -8 (Z-IETD-FMK), and -9 (Z-LEHD-FMK) inhibitors (EMD Bioscience Inc., San Diego, CA, USA) for 40 min before paclitaxel treatment. After 24 h, Hoechst nuclear staining was used to enumerate apoptotic nuclei: small, bright, and condensed nuclei indicate apoptotic cells. Cells were first fixed with ethanol and the DNA-specific dye Hoechst 33342 (Molecular Probes, OR, USA) was added to cells at a final concentration of 0.5 μg/ml. The percentage of apoptotic nuclei was determined by first taking images from four different fields under a fluorescence microscope with 480 nM filter. Each image contained 50–100 cells, and the percentage of apoptotic cells was calculated by dividing the number of cells with bright and small nuclei by the total number of cells.
Measurement of mitochondrial membrane potential with the fluorescent probe JC-1
CRI-619 cells were incubated with or without 40 nM paclitaxel for 24 h and stained with 10 μg/ml JC-1 (Molecular Probes) at room temperature for 10 min. After washing with PBS, cells were analyzed by flow cytometry. Cells with polarized mitochondria presented in the upper-right quadrant of the dot plot due to the formation of JC-1 aggregates (J aggregates), which emit orange color (590 nM) when excited at 488 nM. Cells with depolarized mitochondria emit green color (530 nM) and are visualized in the lower-right quadrant in the dot plot.
Measurement of cell viability by calcein-AM and ethidium homodimer-1 (EthD-1)
CRI-702 cells were treated with or without 40 nM paclitaxel for 24 h and stained with 2 μM calcein-AM and 4 μM EthD-1 solution (Molecular Probe, Eugene, OR, USA) at room temperature for 30 min according to the manufacturer's protocol. The polyanionic dye calcein is well retained within live cells, producing an intense uniform green fluorescence in live cells (ex/em 495/515 nm). EthD-1 enters cells with damaged membranes and undergoes a 40-fold enhancement of fluorescence upon binding to nucleic acids, thereby producing a bright red fluorescence in dead cells (ex/em 495/635 nm).
Abbreviations
- BH:
-
Bcl-2 homologous (domain)
- siRNA:
-
small interfering RNA
References
Green MR (2001) The current status of docetaxel for advanced non-small cell lung cancer. Anticancer Drugs 12 (Suppl 1): S11–S16
Basu A and Haldar S (2003) Identification of a novel Bcl-xL phosphorylation site regulating the sensitivity of taxol- or 2-methoxyestradiol-induced apoptosis. FEBS Lett. 538: 41–47
Basu A, Mohanty S and Sun B (2001) Differential sensitivity of breast cancer cells to tumor necrosis factor-alpha: involvement of protein kinase C. Biochem. Biophys. Res. Commun. 280: 883–891
Burns TF, Fei P, Scata KA, Dicker DT and El-Deiry WS (2003) Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol. Cell. Biol. 23: 5556–5571
Fan S, Cherney B, Reinhold W, Rucker K and O'Connor PM (1998) Disruption of p53 function in immortalized human cells does not affect survival or apoptosis after taxol or vincristine treatment. Clin. Cancer Res. 4: 1047–1054
Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW and Galloway DA (1996) Loss of normal p53 function confers sensitization to taxol by increasing G2/M arrest and apoptosis. Nat. Med. 2: 72–79
Strasser A, Puthalakath H, Bouillet P, Huang DC, O'Connor L, O'Reilly LA, Cullen L, Cory S and Adams JM (2000) The role of bim, a proapoptotic BH3-only member of the Bcl-2 family in cell-death control. Ann. NY Acad. Sci. 917: 541–558
Green DR and Reed JC (1998) Mitochondria and apoptosis. Science 281: 1309–1312
Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM and Strasser A (2002) BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415: 922–996
Cory S and Adams JM (2002) The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2: 647–656
Puthalakath H and Strasser A (2002) Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 9: 505–512
Coultas L, Huang DC, Adams JM and Strasser A (2002) Pro-apoptotic BH3-only Bcl-2 family members in vertebrate model organisms suitable for genetic experimentation. Cell Death Differ. 9: 1163–1166
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB and Korsmeyer SJ (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292: 727–730
Tsujimoto Y (2003) Cell death regulation by the Bcl-2 protein family in the mitochondria. J. Cell. Physiol. 195: 158–167
Srivastava RK, Mi QS, Hardwick JM and Longo DL (1999) Deletion of the loop region of Bcl-2 completely blocks paclitaxel-induced apoptosis. Proc. Natl. Acad. Sci. USA 96: 3775–3780
Srivastava RK, Sasaki CY, Hardwick JM and Longo DL (1999) Bcl-2-mediated drug resistance: inhibition of apoptosis by blocking nuclear factor of activated T lymphocytes (NFAT)-induced Fas ligand transcription. J. Exp. Med. 190: 253–265
Yamamoto K, Ichijo H and Korsmeyer SJ (1999) BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol. Cell. Biol. 19: 8469–8478
Huisman C, Ferreira CG, Broker LE, Rodriguez JA, Smit EF, Postmus PE, Kruyt FA and Giaccone G (2002) Paclitaxel triggers cell death primarily via caspase-independent routes in the non-small cell lung cancer cell line NCI-H460. Clin. Cancer Res. 8: 596–606
Puthalakath H, Huang DC, O'Reilly LA, King SM and Strasser A (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3: 287–296
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM and Strasser A (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738
Broker LE, Huisman C, Ferreira CG, Rodriguez JA, Kruyt FA and Giaccone G (2002) Late activation of apoptotic pathways plays a negligible role in mediating the cytotoxic effects of discodermolide and epothilone B in non-small cell lung cancer cells. Cancer Res. 62: 4081–4088
Song Z, Yao X and Wu M (2003) Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 278: 23130–23140
Yuan SY, Hsu SL, Tsai KJ and Yang CR (2002) Involvement of mitochondrial pathway in Taxol-induced apoptosis of human T24 bladder cancer cells. Urol. Res. 30: 282–288
Blajeski AL, Kottke TJ and Kaufmann SH (2001) A multistep model for paclitaxel-induced apoptosis in human breast cancer cell lines. Exp. Cell Res. 270: 277–288
Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T and Korsmeyer SJ (2001) BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8: 705–711
Wang TH, Wang HS and Soong YK (2000) Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer 88: 2619–2628
Wassmann K and Benezra R (2001) Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev. 11: 83–90
Taylor SS and McKeon F (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89: 727–735
Anand S, Penrhyn-Lowe S and Venkitaraman AR (2003) AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to taxol. Cancer Cell 3: 51–62
Masuda A, Maeno K, Nakagawa T, Saito H and Takahashi T (2003) Association between mitotic spindle checkpoint impairment and susceptibility to the induction of apoptosis by anti-microtubule agents in human lung cancers. Am. J. Pathol. 163: 1109–1116
Ofir R, Seidman R, Rabinski T, Krup M, Yavelsky V, Weinstein Y and Wolfson M (2002) Taxol-induced apoptosis in human SKOV3 ovarian and MCF7 breast carcinoma cells is caspase-3 and caspase-9 independent. Cell Death Differ. 9: 636–642
Hildeman DA, Zhu Y, Mitchell TC, Bouillet P, Strasser A, Kappler J and Marrack P (2002) Activated T cell death invivo mediated by proapoptotic bcl-2 family member bim. Immunity 16: 759–767
Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A and Johnson EM (2001) Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29: 615–628
von Haefen C, Wieder T, Essmann F, Schulze-Osthoff K, Dorken B and Daniel PT (2003) Paclitaxel-induced apoptosis in BJAB cells proceeds via a death receptor-independent, caspases-3/-8-driven mitochondrial amplification loop. Oncogene 22: 2236–2247
Wolf BB, Schuler M, Li W, Eggers-Sedlet B, Lee W, Tailor P, Fitzgerald P, Mills GB and Green DR (2001) Defective cytochrome c-dependent caspase activation in ovarian cancer cell lines due to diminished or absent apoptotic protease activating factor-1 activity. J. Biol. Chem. 276: 34244–34251
Liu JR, Opipari AW, Tan L, Jiang Y, Zhang Y, Tang H and Nunez G (2002) Dysfunctional apoptosome activation in ovarian cancer: implications for chemoresistance. Cancer Res. 62: 924–931
Joseph B, Ekedahl J, Lewensohn R, Marchetti P, Formstecher P and Zhivotovsky B (2001) Defective caspase-3 relocalization in non-small cell lung carcinoma. Oncogene 20: 2877–2888
Joseph B, Marchetti P, Formstecher P, Kroemer G, Lewensohn R and Zhivotovsky B (2002) Mitochondrial dysfunction is an essential step for killing of non-small cell lung carcinomas resistant to conventional treatment. Oncogene 21: 65–77
Egger L, Schneider J, Rheme C, Tapernoux M, Hacki J and Borner C (2003) Serine proteases mediate apoptosis-like cell death and phagocytosis under caspase-inhibiting conditions. Cell Death Differ. 10: 1188–1203
Vande VC, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R and Greenberg AH (2000) BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20: 5454–5468
Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M and Ivashchenko Y (2001) Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 8: 367–376
Arnt CR, Chiorean MV, Heldebrant MP, Gores GJ and Kaufmann SH (2002) Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 insitu. J. Biol. Chem. 277: 44236–44243
McNeish IA, Bell S, McKay T, Tenev T, Marani M and Lemoine NR (2003) Expression of Smac/DIABLO in ovarian carcinoma cells induces apoptosis via a caspase-9-mediated pathway. Exp. Cell Res. 286: 186–198
Yang L, Mashima T, Sato S, Mochizuki M, Sakamoto H, Yamori T, Oh-Hara T and Tsuruo T (2003) Predominant suppression of apoptosome by inhibitor of apoptosis protein in non-small cell lung cancer H460 cells: therapeutic effect of a novel polyarginine-conjugated Smac peptide. Cancer Res. 63: 831–837
O'Connor DS, Wall NR, Porter AC and Altieri DC (2002) A p34(cdc2) survival checkpoint in cancer. Cancer Cell 2: 43–54
O'Reilly LA, Cullen L, Visvader J, Lindeman GJ, Print C, Bath ML, Huang DC and Strasser A (2000) The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am. J. Pathol. 157: 449–461
O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S and Huang DC (1998) Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17: 384–395
Dijkers PF, Medema RH, Lammers JW, Koenderman L and Coffer PJ (2000) Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10: 1201–1204
Gilley J, Coffer PJ and Ham J (2003) FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162: 613–622
Sunters A, Fernandez De Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, Coffer PJ, Medema RH, Coombes RC and Lam EW (2003) FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel treated breast cancer cell lines. J. Biol. Chem.
Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC and Johnson Jr EM. (2003) JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 38: 899–914
Lei K and Davis RJ (2003) JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100: 2432–2437
Weston CR, Balmanno K, Chalmers C, Hadfield K, Molton SA, Ley R, Wagner EF and Cook SJ (2003) Activation of ERK1/2 by deltaRaf-1:ER* represses Bim expression independently of the JNK or PI3K pathways. Oncogene 22: 1281–1293
Ley R, Balmanno K, Hadfield K, Weston C and Cook SJ (2003) Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 278: 18811–18816
Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G and Auberger P (2003) Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22: 6785–6793
Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK and Brugge JS (2003) Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat. Cell Biol. 5: 733–740
Bacus SS, Gudkov AV, Lowe M, Lyass L, Yung Y, Komarov AP, Keyomarsi K, Yarden Y and Seger R (2001) Taxol-induced apoptosis depends on MAP kinase pathways (ERK and p38) and is independent of p53. Oncogene 20: 147–155
Lee LF, Li G, Templeton DJ and Ting JP (1998) Paclitaxel (Taxol)-induced gene expression and cell death are both mediated by the activation of c-Jun NH2-terminal kinase (JNK/SAPK). J. Biol. Chem. 273: 28253–28260
Okano J and Rustgi AK (2001) Paclitaxel induces prolonged activation of the Ras/MEK/ERK pathway independently of activating the programmed cell death machinery. J. Biol. Chem. 276: 19555–19564
Deacon K, Mistry P, Chernoff J, Blank JL and Patel R (2003) p38 mitogen-activated protein kinase mediates cell death and p21-activated kinase mediates cell survival during chemotherapeutic drug-induced mitotic arrest. Mol. Cell. Biol. 14: 2071–2087
Johnson GL and Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298: 1911–1912
Li R, Ruttinger D, Urba WJ, Fox BA and Hu H-M (2004) Targeting and amplification of immune killing of tumor cells by pro-Smac. Int. J. Cancer 109: 85–94
Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM and Bishop JM (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99: 9942–9947
Acknowledgements
We thank Dr. Stanley J Korsmeyer for providing the retroviral vectors, Dr. Walter J Urba for the critical reading of this manuscript, Dr. Bernard A Fox for his support, and Mr. Dan Haley for the cell sorting. This study was supported by grants from the Chiles Foundation, the Providence Portland Medical Foundation, the MJ Murdock Charitable Trust, and Cell Genesis Corporation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Li, R., Moudgil, T., Ross, H. et al. Apoptosis of non-small-cell lung cancer cell lines after paclitaxel treatment involves the BH3-only proapoptotic protein Bim. Cell Death Differ 12, 292–303 (2005). https://doi.org/10.1038/sj.cdd.4401554
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401554
Keywords
This article is cited by
-
Acetylation of FOXO1 activates Bim expression involved in CVB3 induced cardiomyocyte apoptosis
Apoptosis (2023)
-
Utidelone inhibits growth of colorectal cancer cells through ROS/JNK signaling pathway
Cell Death & Disease (2021)
-
PRKCQ inhibition enhances chemosensitivity of triple-negative breast cancer by regulating Bim
Breast Cancer Research (2020)
-
Penfluridol overcomes paclitaxel resistance in metastatic breast cancer
Scientific Reports (2019)
-
Resistance to anti-microtubule drug-induced cell death is determined by regulation of BimEL expression
Oncogene (2019)