Abstract
Cancer develops when molecular pathways that control the fine balance between proliferation, differentiation, autophagy and cell death undergo genetic deregulation. The prospects for further substantial advances in the management of colorectal cancer reside in a systematic genetic and functional dissection of these pathways in tumor cells. In an effort to evaluate the impact of p38 signaling on colorectal cancer cell fate, we treated HT29, Caco2, Hct116, LS174T and SW480 cell lines with the inhibitor SB202190 specific for p38α/β kinases. We report that p38α is required for colorectal cancer cell homeostasis as the inhibition of its kinase function by pharmacological blockade or genetic inactivation causes cell cycle arrest, autophagy and cell death in a cell type-specific manner. Deficiency of p38α activity induces a tissue-restricted upregulation of the GABARAP gene, an essential component of autophagic vacuoles and autophagosomes, whereas simultaneous inhibition of autophagy significantly increases cell death by triggering apoptosis. These data identify p38α as a central mediator of colorectal cancer cell homeostasis and establish a rationale for the evaluation of the pharmacological manipulation of the p38α pathway in the treatment of colorectal cancer.
Similar content being viewed by others
Main
Colorectal cancer is a major health concern, with more than 1 000 000 new cases and 500 000 deaths expected worldwide per year.1 Prognostic evaluation is currently based on histological appearance, and there are no molecular markers internationally recognized as standard predictor factors. The conventional therapy involving surgery and adjuvant therapy seems to give rise to improvements in progression-free and overall survival. Nevertheless about 50% of patients die within 5 years owing to metastasis or recurrent disease.2 The prospects for further substantial advances in the management of colorectal cancer reside in a systematic genetic and functional dissection of cell cycle and cell death regulatory pathways in tumor cells in order to identify differential cellular effects of agents that may have a direct impact on cancer therapy.
During the last decade, a number of deacetylase inhibitors (DI) have been identified. These DI induce tumor cells to undergo growth arrest, differentiation, and/or apoptosis in culture and in animal models, at doses that seem to be non-toxic and appear to be selective. Butyrate, a DI that is naturally formed in the human colon, is able to reduce the size and the number of tumors in rat models of bowel cancer.3 In vitro, sodium butyrate (NaB) is a potent differentiating agent for several colorectal cancer cell lines (CRCs).4, 5, 6 NaB-mediated cell cycle withdrawal of CRCs appears to be dependent on acetylation of histones and consequent changes in transcription, requiring continuous protein synthesis and the expression of p21.7 It has recently been shown that 1 mM NaB is the best working concentration to activate the differentiation program in CRCs without triggering apoptosis, whereas 5 mM NaB is sufficient to induce a p53-independent apoptotic cell death by activating a pathway involving p38, PPARγ and caspases.6, 8
Several intracellular signaling cascades have been implicated in the regulation of the proliferation-differentiation balance in enterocytes by the coordinated expression of the genome in response to environmental cues. During enterocyte differentiation of CRCs the ERK pathway is switched off, whereas PI3K class I and p38 activities are increased and essential for full differentiation.5, 9 It has been suggested that p38α could activate the intestinal-specific homeobox transcription factor CDX2 probably by phosphorylation.10 This mechanism resembles that described for p38α/β in muscle cells, where they phosphorylate the transcription factor MEF2 and the SWI/SNF subunit BAF60 to remodel chromatin and activate tissue-specific transcription.11, 12 Interestingly, chromatin remodeling complex dysfunction has been described in human colorectal cancer cells.13
The p38 pathway is often activated by stressful stimuli and cytokines leading to diverse cell type-specific responses, such as cell survival or apoptosis. In non-transformed cells, p38α is required for inhibition of proliferation induced by cell-to-cell contact, as happens in primary cultures of human fibroblasts.14 In cancer cells, p38α/β are required for breast and colorectal cancer cell migration and metastasis formation in mice.15, 16 However, over-activation of p38 by a constitutively active form of its upstream kinase (MKK6EE) eliminates the tumor-prone phenotype of mice obtained by intercrossing transgenic mice with breast-restricted expression of Wip1, the p53-induced phosphatase targeting p38 and the oncogene ErbB2.17 Moreover, p38α/β contribute to TGFβ-dependent tumor progression by inducing epithelial to mesenchymal transdifferentiation and cell migration,18 and to rat chondrosarcoma cell proliferation.19
In an effort to elucidate the impact of p38 in the molecular mechanisms regulating CRC fate, we analyzed gene expression profile of HT29 cells growing logarithmically, confluent in culture and induced to differentiate. Five genes were considered, three of them were cell cycle-related genes: cyclin E is expressed during the late G1 phase of the cell cycle until the end of the S-phase, cyclin A starts to accumulate during S phase and is abruptly destroyed before metaphase, and, finally, p21 is involved in the control of G1/S and G2/M transitions. The other two genes codify for tissue-specific proteins: sucrose isomaltase (SI) is an intestinal disaccharidase expressed in the brush border of mature enterocytes; villin is a cytoskeletal protein specific for microvilli.
At the onset of differentiation, the pharmacological blockade of the p38 cascade inhibits the expression of p21, SI and villin confirming the role of this pathway in the differentiation program of distinct cell types.9, 10, 11, 12 Surprisingly, we discovered that p38α is required for CRC proliferation and survival as pharmacological blockade or genetic depletion of its kinase activity induced growth arrest, autophagy and cell death in a cell type-specific manner. Interestingly, the inhibition of the autophagic activity of these cells induces a dramatic increase in cell death by triggering apoptosis.
Results
Pharmacological blockade of p38 affects proliferation and differentiation of HT29 cells
To characterize the molecular mechanisms involving the p38 pathway in the control of colorectal cancer cell fate, we employed the human HT29 cell line that recapitulates large bowel cell differentiation in vitro. The effect of p38 inhibition by the specific compound SB202190 (SB) was evaluated by analyzing the expression of cyclin E, cyclin A, p21, SI and villin transcripts in HT29 cells growing logarithmically, confluent in culture or induced to differentiate with NaB 1 mM for 96 h (Figure 1a). SB addition to logarithmically growing cultures caused a significant reduction of cyclin E expression already within the first 24 h, whereas it downregulated cyclin A transcription only after 72 h of treatment (Figure 1b). Moreover, despite the significant reduction in the proliferation rate, the levels of p21 were consistently lower than those of untreated cells (Figure 1b). Conversely, in confluent cultures, SB only slightly affected p21 expression induced by cell-to-cell contact (Figure 1b). In differentiating HT29 cells, p38 blockade counteracted the effect of NaB on the silencing of cyclins, induction of p21 and expression of enterocyte-specific markers (Figure 1b). Collectively these data suggested that the p38 pathway is involved in the transcription control of both cell cycle and tissue-specific genes.
Inhibition of p38 by SB202190 modifies the expression profile of HT29 cells growing logarithmically, confluent in culture and induced to differentiate. (a) Cells were plated at different density in either the absence or presence of NaB and SB202190 (SB) and cultured for up to 96 h. (b) Relative levels of expression of the indicated genes were calculated by GeneScan and normalized on GAPDH signal (see Supplementary Figure 1). (c) For each set of primers a standard curve was constructed using serial dilutions of cDNA to define the dynamic range and calculate the amplification efficiency. Relative PCR levels were estimated by normalization on the GAPDH fluorescent signal obtained by the amplification of 1/10 cDNA dilution (see asterisk)
p38 blockade induces autophagic cell death in HT29 cells
Very surprisingly, p38 blockade in HT29 cells induced the formation of large cytoplasmic vacuoles regardless of culture conditions (Figure 2), whereas PD98059 and LY294002, two inhibitors specific for MEK1 and PI3K kinases, respectively, did not (data not shown). They appeared approximately 2 h after SB addition, then became larger reaching the maximum diameter at 24 h (19.3±10.6 μM) (Figure 2a). The percentage of cells presenting vacuoles was time dependent as well. At 24 h more than 90% of cells presented at least one vacuole filling up most of the cytoplasmic compartment (Figures 2 and 4a).
p38 blockade induces the formation of macrovacuoles in HT29 cells. HT29 cells were cultured in either the absence or presence of NaB 1 mM or SB202190 (SB) for 24 h. (a) Living cells observed and photographed with a phase-contrast microscope. (b) Cells fixed and stained with PAS showing nucleolated nuclei and granular cytoplasms (untreated); hyperchromatic nuclei and single intracytoplasmic macrovacuole (SB); nucleolated nuclei and granular cytoplasms with PAS positive deposits (NaB). SB-induced macrovacuoles show a dot-like staining (inset). Original magnification × 400
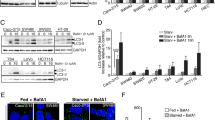
p38 blockade induces autophagic cell death in HT29 cells, whereas inhibition of SB202190-dependent autophagy triggers apoptosis. HT29 cells were cultured in the absence or presence of SB202190 (SB) or a combination of SB202190 and 3-methyiladenine (3MA) or SB202190 and puromycin (Puro) for the indicated periods of time. (a) The percentage of autophagic cells was estimated by phase-contrast microscope and then verified by electron microscope evaluation. (b) Time course of GABARAP gene expression. Relative levels of GABARAP expression were measured by GeneScan evaluation of fluorescent amplifications and normalized on GAPDH signals. (c–f) HT29 cells were collected at the indicated periods of time and mixed with trypan blue solution or a dye mix containing DAPI and ethidium bromide. (c) Relative viability (%) was estimated by calculating the ratio between viable cells in treated (SB, SB+3MA, SB+Puro) versus untreated (−) HT29. (d) Relative cell death (%) represents the ratio between dead cells in treated versus untreated HT29. (e) HT29 cells stained with DAPI+ethidium bromide and analyzed by fluorescent microscope ( × 400). (f) The ratio between apoptotic and necrotic dead cells was calculated and reported as a percentage of the total number of cell counted (g) Genomic DNA was extracted from HT29 cells and loaded into a 1% agarose gel
The observation that SB treatment caused growth inhibition and the formation of intracellular vacuoles, prompted us to investigate their nature. To this end we employed different cytological methods such as periodic-acid Schiff (PAS), Papanicolaou, Sudan black B and Oil red O. Among them only PAS demonstrated a dot-like staining within vacuoles suggesting the presence of glycoproteins (Figure 2b). Electron microscopy showed that SB-treated cells were roundly shaped and displayed a dark cytoplasm containing a heterochromatic nucleus. Numerous large vacuoles filled up with dense material, autophagic vacuoles containing cell organelle debris and double-membrane autophagosomes were recognized (Figure 3). In untreated HT29 cells no molecular features of autophagy were detected (data not shown). Quite different was the morphology of fully differentiated HT29 cells, which appeared elongated, and displayed euchromatic nuclei and electron clear cytoplasms with numerous mitochondria and a few small empty vacuoles. Moreover, circular structures coated with regular microvilli, often located at the apical cellular surface (intracellular lumens, IL) or between the cells (intercellular cysts, IC), were observed (Figure 3). In HT29 cells either untreated or treated with SB no IL or IC were detected (Figure 3 and data not shown).
Electron microscopy of HT29 cells cultured in the presence of NaB 1 mM (96 h) or SB202190 (SB; 24 h). (a) NaB-treated cells showing euchromatic nuclei and cytoplasms containing a few vacuoles (arrowhead) and numerous mitochondria (m). Microvilli (arrow) are present on the external plasma membrane. (b) An IC coated by regular microvilli (asterisk). (c, d) SB-treated cells show heterochromatic nuclei (n) and dark cytoplasms filled up with large vacuoles containing dense material (asterisk), autophagic vacuoles containing debris of a mitochondrium (m) (d, inset), double-membrane autophagosomes (c, white arrow) and lipidic droplets (c, arrow). Scale bar: (a and c) 0.7 μm; (b) 0.4 μm; (d) 0.57 μm
The detection of these ultrastructural findings, defined as the ‘golden standard’ for assessing autophagic activity,20 clearly indicated that pharmacological blockade of p38 induced an autophagic response in HT29 cells. Further evidence was provided by the pretreatment with 3-methyladenine (3MA) or with the inhibitor of translation puromycin, two well-known inhibitors of the autophagic process, that severely impaired SB-dependent induction of autophagy in HT29 cells (Figure 4a). To better characterize this process, we evaluated the expression of the GABARAP gene, one of the human homologs of the yeast ATG8 gene. Atg8 is a member of a novel ubiquitin-like protein family and an essential component of the autophagic machinery. In yeast, it is upregulated by rapamycin-dependent Tor inhibition and nitrogen starvation.21 In HT29 cells, GABARAP expression levels were significantly augmented by p38 blockade and correlated with autophagic vacuoles and autophagosome formation (Figure 4a and b), whereas rapamycin failed to induce both autophagy and GABARAP upregulation in these cells (data not shown). Interestingly, 3MA failed to impair the SB-mediated induction of GABARAP expression (Figure 4b). This is noteworthy as 3MA prevents autophagy at an early stage by inhibiting PI3K class III, which interacts with Atg6/Beclin-1 to promote the nucleation of autophagic vesicles.22, 23 Our data suggest that the p38 control of GABARAP transcription is independent from this first step.
In addition to the above-mentioned growth suppression of HT29 cells (Figure 1a), SB exposure significantly reduced the number of viable cells compared to the untreated control cells (relative viability) by triggering cell death (Figure 4c and d). Non-viable SB-treated cells showed the characteristic features of necrotic-like cells. In fact, they took up ethidium bromide, a marker of plasma membrane disruption, but maintained the nucleus intact until late stages, and showed only a partial chromatin condensation without a significant increase in DNA fragmentation (Figure 4e–g). The coexistence of autophagy with features of non-apoptotic death is considered as an hallmark of autophagic cell death or type II programmed cell death.20, 24 Notably, pharmacological blockade of SB-dependent autophagy not only failed to increase viability, but it even increased cell death by inducing a switch from autophagic cell death to apoptosis. In fact, cells pretreated with 3MA and then exposed to SB showed cell shrinkage, severe chromatin condensation, nuclear fragmentation and DNA degradation comparable to cells exposed to NaB 5 mM (Figure 4e–g).
SB-dependent induction of autophagy relies only upon p38α blockade
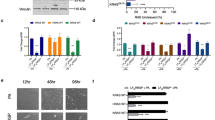
As SB specifically inhibits p38 α and β gene products, we investigated whether both isoforms were synthesized in HT29 cells. It was previously reported that Caco2 cells exhibit distinct profiles of p38 isoform expression depending on the differentiation status. Undifferentiated cells produce p38α, β and γ, whereas differentiated cells synthesize p38α, γ and δ.25 Conversely, the expression profile of p38 isoforms in HT29 cells remained unchanged regardless of the differentiation status. In fact, the cells expressed p38α, γ and δ despite NaB treatment (Figure 5a and b). The absence of p38β transcripts suggested that SB-mediated autophagic response relies only upon p38α blockade. To test this hypothesis, we used siRNA gene silencing targeting p38 α and β gene in HT29 cells (Figure 5e). The siRNA against p38α reduced the target mRNA and protein levels by 70–80%, whereas the siRNA against the β isoform did not affect nor p38α mRNA neither total p38 protein expression in accordance with the absence of p38β in these cells (Figure 5c). Evaluation of the autophagic activity and GABARAP expression in siRNA-transfected cells clearly indicated that transient genetic ablation of p38α was able to induce autophagy in HT29 colorectal cancer cells (Figure 5d).
SB202190-dependent induction of autophagy relies only upon p38α blockade. (a, b) HT29 cells cultured in the absence or presence of NaB 1 mM (48 h) were characterized for the expression of p38 α, β, γ and δ isoforms. (a) Multiplex RT-PCR analysis was followed by separate PCR confirmation for each transcript. (b) Relative levels of expression of p38 α and β isoforms were measured by GeneScan evaluation of fluorescent amplifications and normalized on GAPDH signals. (c–e) HT29 cells were transfected with siRNA targeting p38α and β genes. After 24 h RNA was extracted, and the expression of p38 isoforms was evaluated by real-time PCR (c, upper panel). At 48 h post-transfection cell lysates were probed for p38 expression by immunoblot (c, lower panel), and expression of GABARAP was measured by GeneScan evaluation of fluorescent amplifications and normalized on GAPDH signals (d, lower panel). The percentage of autophagic cells was estimated by phase-contrast microscope (e) and then verified by electron microscopy evaluation (d, upper panel)
Colorectal cancer-specific induction of autophagic cell death by p38α inhibition
To get further insight into the involvement of p38α in the autophagic response, we extended our investigation to Caco2, SW480, LS174T and Hct116 CRCs (Tables 1 and 2). Cytological and morphological evaluation confirmed the SB-mediated induction of growth arrest and autophagic cell death in all CRC lines (Figures 6a–c). Consistently, SB-mediated upregulation of GABARAP was detected in all cell lines tested (Figure 6e). Pharmacological blockade of autophagy drastically increased cell death by promoting a switch from autophagic death to apoptosis (Figure 6c, and data not shown).
SB202190 induces autophagic cell death in colorectal cancer cells in a cell type-specific manner. (a–c) SW480, CaCo2 and Hct116 cells were cultured in the absence or presence of SB202190 (SB) or a combination of SB202190 and 3-methyiladenine (3MA) for the indicated period of time. Cells were suspended in a trypan blue solution or in a dye mix containing DAPI and ethidium bromide. (a) Relative viability (%) was estimated by calculating the ratio between viable cells in treated (SB, SB+3MA) versus untreated (−) HT29. (b) The percentage of autophagic cells was estimated by phase-contrast microscopy and then verified by electron microscope evaluation. (c) Relative cell death (%) represents the ratio between dead cells in treated versus untreated HT29. (d) Equal number of HeLa, hFSF, Hep3B and HEK293 cells were seeded and cultured in the absence or presence of SB202190 for 24 h. Cells were scored for autophagic activity (data not shown), and viability by trypan blue staining (viable cells/total number of cells). (e, f) SW480, CaCo2, Hct116, HeLa, hFSF, Hep3B and HEK293 cells were cultured in the absence or presence of SB202190 for 24 h. Relative abundance of GABARAP (e) or p38 α and β (f) transcripts in the indicated cell lines were evaluated by GeneScan estimation of fluorescent RT-PCR products and normalization on GAPDH
To investigate whether p38α inhibition induced type II cell death in a cell type-specific manner, Hep3B liver carcinoma cells, HeLa cervical carcinoma cells, HEK293 embryonic kidney cells, and human fetal skin fibroblasts (hFSF) were exposed to SB for 24 h (Figure 6d and Table 2). Pharmacological inhibition of p38 failed to induce cell cycle arrest, autophagy and cell death, and it did not promote GABARAP upregulation in any of the cell lines tested (Figure 6d and e, and data not shown).
The analysis of the relative abundance of p38 α and β transcripts in all cell lines studied, suggested that the cell type-specific effect caused by SB is independent of the expression of p38β and relies only upon the presence of p38α in CRCs (Figure 6f).
Moreover, SB treatment failed to cause autophagic cell death in C3H10T1/2 and NIH3T3 mouse fibroblasts, C2C12 mouse myoblasts and human RD rhabdomyosarcoma cells (Table 2). All together, these data suggest that p38α blockade induces growth inhibition, autophagy and cell death in CRCs in a cell type-specific manner.
Discussion
We demonstrated that p38α is one of the essential kinases for colorectal cancer cells fate and provided evidences indicating that p38α blockade causes growth arrest and autophagic cell death. In CRCs, p38α is required for both cell cycle progression and NaB-dependent induction of cell differentiation. Furthermore, our experiments reveal that p38α is required for survival of CRCs growing logarithmically, confluent in culture or induced to differentiate. In fact, p38α blockade induces an autophagic response in these cells associated to growth suppression and cell death in a p53- and p21-independent manner.
The involvement of p38 activity in the negative control of autophagy has been evoked in human liver response to cellular hydration state, but not in mouse myotube response to amino-acid starvation.26 In yeasts, Hog1, the homolog of p38, plays a role in the stabilization machinery of nitrogen deprivation-induced autophagy during ambient osmolarity changes.27 In logarithmically growing CRCs, p38α inhibition induces a cell type-specific upregulation of the human homolog of Atg8 (GABARAP), suggesting that p38α is required to exert a negative control on the expression of this gene. In yeasts, the Atg8 gene is upregulated by rapamycin-dependent Tor inhibition, and its protein product is an essential component of the autophagic vescicle formation/size expansion machinery.21 In CRCs, rapamycin failed to cause both autophagy and upregulation of GABARAP expression. To date, three Atg8 homologs have been identified in mammalian cells: GATE-16, GABARAP and MAP1LC3. All three proteins appear to be modified with lipids in the same manner described in yeasts, and have been shown to localize to the autophagosome.28
Very interestingly, pretreatment with 3MA or puromycin reduces the number and size of vacuoles together with the percentage of autophagic cells in SB-treated CRCs, whereas the induction of GABARAP expression remains unaffected. 3MA controls the activity of the PI3K class III enzyme which is important in the early stages of autophagic vescicle formation by inducing the assembly of autophagosome precursors.22, 23 In yeast, PI3K/Vps34 forms a multiprotein complex including also Atg6 and Atg14. In humans, the mammalian homolog of Atg6, named Beclin-1, has been found mutated in cancer cells revealing its causative role for autophagy deficiency in cancerogenesis.20
Autophagy is a cellular process that can finally consent cell survival in stress conditions or trigger type II cell death depending on the cell type and specific stimuli.24, 28 Recent work indicates that several intracellular mediators of autophagy are deregulated during malignant transformation, with a consequent reduction of the autophagic activity.20, 24, 29 Cancer cell lines often have lower autophagic capacity than their normal counterparts and fail to respond to serum/amino-acid deprivation or high cell density. A decrease in autophagic capacity has been also observed during animal experimental carcinogenesis.20 Another point underscoring the relevance of autophagy in cancer cells derives from the observation that some malignant cell types respond to chemotherapy or radiation by triggering autophagy, indicating the potential utility of autophagic cell death induction in cancer therapy.30 Treatment with rapamycin, temozolomide or arsenic trioxide of malignant glioma cells induces an autophagic cell death response.31, 32, 33 However that's not always the case. In fact, characterization of the effects of two DNA-damaging agents, such as temozolide and etoposide, suggests that these chemotherapy compounds induce an autophagy-associated ATP surge that protects cells contributing to drug resistance of malignant glioma.34 Moreover, autophagy could represent a defense mechanism against sulforaphane-induced apoptosis in prostate cancer cells and against DNA-damaging agents in breast cancer.35, 36 It has been recently shown that when cells with defects in apoptosis are subjected to metabolic stress, they activate autophagy to survive. Inhibition of autophagy triggers a necrotic pathway leading to cell death. In solid tumors, this necrosis is associated with inflammation, which creates a microenvironment favoring survival and increased proliferation of a small population of damaged cells. The final effect of this process is the enhancement of tumor growth.37 Thus, it seems that the role of autophagy in triggering survival or death in response to cancer therapies may vary depending on different cell types and treatments.
According with our results, it seems that p38α blockade in CRCs inhibits a survival pathway leading to autophagy, which probably initially represents a defense mechanism to survive. Along the treatment, these autophagic cells die with features of non-apoptotic death indicating the occurrence of autophagic cell death or type II programmed cell death. Although the increased percentage of dead cells caused by 3MA-dependent inhibition of autophagy could appear in conflict with this hypothesis, but these cells actually showed distinctive traits of apoptosis. To explain these data we propose a model suggesting that autophagy represents a survival response to p38α blockade, but prolonged inactivation of the kinase leads to cell death. Inhibition of autophagy breaks this process and triggers an apoptotic response. The effect of SB treatment in CRCs recalls that of arsenic trioxide in glioma cells. In fact, the addition of an autophagy inhibitor enhances the antitumor effect through the switch between autophagic to apoptotic cell death.33 In accordance with this hypothesis, removal of SB within the first 48 h of treatment induces a significant time-dependent reduction in vacuole number and size together with a slow re-entry in the cell cycle with a dubling time of about 80 h (Comes and Simone, unpublished results).
Our experiments reveal that SB induces autophagic cell death in a cell type-specific manner as normal cells and cancer cells of different origin fail to respond to p38α blockade. Therefore, our findings establish a rationale for the evaluation of p38α-specific inhibition as potential therapeutic tool, alone or in combination with other compounds, in the treatment of colorectal cancer. In fact, as reported in Table 1, the CRC lines used in these study display significant differences both in genotype and phenotype. None of the molecular characteristics of these cells seem to be correlated with the common autophagic cell death response to SB. Moreover, BALB/c mice treated daily by intraperitoneal injection of SB for nine days survived without body and colon weight changes. It is noteworthy that the colon of these mice showed normal architecture at the histological analysis.38 Furthermore, a novel strategy for local delivery of SB has been developed to treat renal fibrosis,39 indicating that pharmacological manipulation of p38 is emerging as a potential therapeutic tool for a number of human diseases. In this sense, our study might have an even broader interest. In conclusion, the experimental model we have described can represent a useful system to study human autophagy in vitro.
Materials and Methods
Reagents
SB202190 (10 μM), PD98059 (20 μM), LY294002 (25 μM), rapamycin (100 nM) and NaB (1–5 mM) were purchased from Calbiochem; puromycin (10 μM), 3-methyladenine (10 mM), Sudan black B, Oil red O, trypan blue, DAPI and ethidium bromide were purchased from Sigma.
Cell culture and microscopic quantitation of proliferation, viability and cell death
HT29, SW480, Hct116 and Caco2 cells were cultured as described.40 LS174T, Hep3B, HeLa, HEK293, C2C12, CH310T1/2, NIH3T3 and RD cells were maintained in DMEM with 10% FBS. hFSF were isolated and cultured as described.40 The proliferation, viability and cell death of the reported cell lines were scored by cell counting. Briefly, at the indicated time points the supernatants (containing dead/floating cells) were collected, and the remaining adherent cells were detached with Trypsin/EDTA. After that, both cell suspensions were mixed and cell pellets were obtained. Cell pellets were suspended in PBS 1 × and then 10 μl were mixed with equal volume of 0.01% trypan blue solution. The mixture was then transferred to the hemacytometer. Viable cells (unstained cells) and dead cells (stained cells) were counted with a phase-contrast microscope ( × 40). To score the proliferation rate the entire counting grid was analyzed, whereas to evaluate viability 200 cells were counted. To determine the percentage of cells dying for apoptotic or non-apoptotic cell death, 25 μl of cell suspensions were mixed with 1 μl of dye mix containing DAPI (1 μg/ml) and ethidium bromide (100 μg/m). The mixture was placed on a microscope slide and covered with a 22 mm2 coverslip. The slides were examined with the ‘Zeiss Axioskop 2 plus’ fluorescent microscope ( × 400). For each sample 200 cells were counted and recorded as V (viable cells), NVN (non-viable cells with normal nuclei) and NVA (non-viable cells with apoptotic nuclei). The % of apoptotic versus non-apoptotic dead cells was calculated (% apoptotic cells=100 × NVA/(VA+NVN+NVA); % non-apoptotic cells=100 × NVN/(VA+NVN+NVA)).
To avoid possible influence of cell density on cell growth and survival, every set of experiments (at least three replicates) was performed by seeding cells at different densities: from 50 × 103 to 500 × 103 cells in six-well dishes and from 500 × 103 to 1000 × 103 in 60-mm-diameter dishes. The data shown in the ‘Results’ section are representative of three or more independent sets of experiments.
DNA fragmentation assay
The supernatants were mixed with the remaining adherent cells detached with Trypsin/EDTA, and cell pellets were suspended in hypotonic lysis buffer. After centrifugation at 14 000 r.p.m. for 20 min, proteinase K and then RNAsi A, was added. Samples were subjected to double phenol/chloroform extraction, DNA precipitation, suspension in TE and loaded into 1% agarose gel.
RNA interference
HT29 cells were seeded at 1500 × 103 cells/well in six-well dishes. Twenty-four hours after seeding, cells were transfected with 200 pmol of siGenome SMARTpool reagent from DHARMACON (M-003512-05-0005) containing four siRNA duplexes direct against p38α or p38β open-reading frame using 10 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
RNA extraction and cDNA synthesis
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instruction. To avoid possible DNA contaminations, RNA was treated with DNAase-1 (Ambion, Austin, TX, USA). RNA purity was also checked by spectrophotometer and RNA integrity by examination on agarose gel electrophoresis. In every experiment, one to four micrograms of total RNA were retro-transcribed using High Capacity DNA Archive Kit (Applied Biosystem, Foster City, CA, USA) and following the manufacturer's instruction.
Semiquantitative RT-PCR
RT-PCR was performed using specific primers whose sequence is available on request. Standard curves were performed using serial dilutions of cDNA to define the dynamic range and calculate the amplification efficiency (see Figure 1). The PCR conditions were the following: 5 min at 94°C for one cycle; 30 s at 94°C, 30 s at 59–62°C and 1 min at 68°C for 22–28 cycles. To quantify the relative abundance of gene-specific transcripts, fluorescent-labelled PCR products were analyzed on an ABI310 sequencer (Applied Biosystems, Foster City, CA, USA) with GeneScan software following the manufacturer's instruction, and normalized to GAPDH transcripts (see Supplementary Figure 1). Multiplex RT-PCR was performed with Quiagen Multiplex PCR Master Mix using specific primers described previously.25
Quantitative real-time PCR
To detect mRNA expression levels for the p38 α and β genes, primers were designed using Primer Express software spanning intron/exon boundaries whenever possible. PCR assays were performed in 96-well optical reaction plates using the ABI 7500HT machine (Applied Biosystem, Foster City, CA, USA). PCR assays were conducted in triplicate wells for each sample. The following reaction mixture per well was used: 10 μl Power Syber Green (Applied Biosystem, Foster City, CA, USA), 2.4 μl of primers at the final concentration of 150 nM, 4.6 μl RNAase free water, 3 μl cDNA. For all experiments the following PCR conditions were used: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s then at 60°C for 60 s. Quantitative normalization of cDNA in each sample were performed using cyclophillin mRNA as internal control. Relative quantification was carried out using the ΔΔCT method. Mock-transfected cells were used as calibrator to determine relative expression level.
Immunoblot assay
Immunoblots were performed according to the Cell Signaling instructions. Briefly, cells were homogenized by sonication in 1 × SDS sample buffer (62.5 mM Tris-HCl pH 6.8, 2% w/v SDS, 10% glycerol and 50 mM DTT). 50 μg of protein extract from each sample were electrophoresed in a 10% SDS-polyacrylamide gel and transferred to Hybond-P PVDF membrane (Amersham Biosciences, Piscataway, NJ, USA). The blots were blocked 1 h at RT in blocking buffer (1 × TBS, 0.1% Tween-20 with 5% w/v non-fat dry milk) and washed with TBS/T. The membranes were incubated with anti-p38 antibody (Cell Signaling; 1 : 1000) in 10 ml primary antibody dilution buffer (1 × TBS, 0.1% Tween-20 with 5% BSA) overnight at 4°C. After that, the blots were extensively washed and incubated with the anti-rabbit secondary horseradish peroxidase conjugate antibody (1:20 000; Amersham Biosciences, Piscataway, NJ, USA) in blocking buffer for 45 min, then washed and developed with the ECL chemiluminescence reagent (Amersham Biosciences, Piscataway, NJ, USA) as directed by the manufacturer.
Morphological evaluation
Numerous slides with a monolayer cells were either rapidly fixed in 95% ethyl alcohol for a minimum of 15 min (before staining with Papanicolaou and PAS) or unfixed (before staining for lipids with Sudan black B, Oil red O). Slides were examined by optic microscopy (Nikon Eclipse 80i).
Measurement of vacuole size
Digital images of living vacuolated cells were imported on Adobe Photoshop software, version 7.0. The diameter of each vacuole in 50 randomly selected cells was measured and expressed in a pixel scale. The length of one grid in a micro-grid slide was also measured in a pixel scale. The diameter of each vacuole on a pixel scale was converted into micrometers based on the pixels of the standard grid length. The resolution of all the images was kept constant during the whole procedure.
Electron microscopy
Cells were fixed in 3% GTA in phosphate buffer 0.1 M for 2 h, washed in the same buffer and then post-fixed in 1% OsO4 at 4°C. Afterwards, cells were scraped with a rubber bar, dehydrated in graded ethanols, and embedded in Epon 812. Ultrathin sections (60 nm) were cut with a diamond knife on a LKB-V ultratome, stained with uranyl acetate followed by lead cytrate and examined under a ‘Zeiss 9A’ electron microscope. To score autophagy 100 cells for each sample were counted.
Abbreviations
- DI:
-
deacetylase inhibitors
- NaB:
-
sodium butyrate
- CRCs:
-
colorectal cancer cell lines
- SI:
-
sucrose isomaltase
- SB:
-
SB202190
- PAS:
-
periodic-acid Schiff
- IL:
-
intracellular lumens
- IC:
-
intercellular cysts
- 3MA:
-
3-methyladenine
- hFSF:
-
human fetal skin fibroblasts
References
Samoha S, Arber N . Cyclooxygenase-2 inhibition prevents colorectal cancer: from the bench to the bed side. Oncology 2005; 69 (Suppl 1): 33–37.
McGartland LP, Mulcahy MF, Benson III AB . Pre- and postoperative adjuvant therapy for locally advanced rectal cancer. Clin Adv Hematol Oncol 2004; 2: 806–814.
Medina V, Afonso JJ, Alvarez-Arquelles H, Hernandez C, Gonzalez F . Sodium butyrate inhibits carcinoma development in a 1,2-dimethylhydrazine-induced rat colon cancer. J Parenter Enteral Nutr 1988; 22: 14–17.
Van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet 2004; 36: 277–282.
Luongo D, Mazzarella G, Della RF, Maurano F, Rossi M . Down-regulation of ERK1 and ERK2 activity during differentiation of the intestinal cell line HT-29. Mol Cell Biochem 2002; 231: 43–50.
Orchel A, Dzierzewicz Z, Parfiniewicz B, Weglarz L, Wilczok T . Butyrate-induced differentiation of colon cancer cells is PKC and JNK dependent. Dig Dis Sci 2005; 50: 490–498.
Archer SY, Johnson J, Kim HJ, Ma Q, Mou H, Daesety V et al. The histone deacetylase inhibitor butyrate downregulates cyclin B1 gene expression via a p21/WAF-1-dependent mechanism in human colon cancer cells. Am J Physiol Gastrointest Liver Physiol 2005; 289: G696–G703.
Schwab M, Reynders V, Ulrich S, Zahn N, Stein J, Schroder O . PPARgamma is a key target of butyrate-induced caspase-3 activation in the colorectal cancer cell line Caco-2. Apoptosis 2006; 11: 1801–1811.
Laprise P, Chailler P, Houde M, Beaulieu JF, Boucher MJ, Rivard N . Phosphatidylinositol 3-Kinase controls human intestinal epithelial cell differentiation by promoting adherens junction assembly and p38 MAPK activation. J Biol Chem 2002; 277: 8226–8234.
Houde M, Laprise P, Jean D, Blais M, Asselin C, Rivard N . Intestinal epithelial cell differentiation involves activation of p38 mitogen-activated protein kinase that regulates the homeobox transcription factor CDX2. J Biol Chem 2001; 276: 21885–21894.
Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL . p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 2004; 36: 738–743.
Simone C . SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. J Cell Physiol 2006; 207: 309–314.
Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene 1999; 18: 3422–3426.
Faust D, Dolado I, Quadrado A, Oesch F, Weiss C, Nebrada AR et al. p38alpha MAPK is required for contact inhibition. Oncogene 2005; 24: 7941–7945.
Surez-Cuervo C, Merrell MA, Watson L, Harris KW, Rosenthal EL, Vaananen HK et al. Breast cancer cells with inhibition of p38alpha have decreased MMP-9 activity and exhibit decreased bone metastasis in mice. Clin Exp Metastasis 2004; 21: 525–533.
Gout S, Morin C, Haule F, Huot J . Death receptor-3, a new E-Selectin counter-receptor that confers migration and survival advantages to colon carcinoma cells by triggering p38 and ERK MAPK activation. Cancer Res 2006; 66: 9117–9124.
Demidov ON, Kek C, Shreeram S, Timoffev O, Fornace AJ, Appella E et al. The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene 2006; (E-pub ahead of print).
Bakin AV, Rinehart C, Tomlinson AK, Harteaga CL . p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci 2002; 115: 3193–3206.
Halawani D, Mondeh R, Stanton LA, Beier F . p38 MAP kinase signaling is necessary for rat chondrosarcoma cell proliferation. Oncogene 2004; 23: 3726–3731.
Gozuacik D, Kimchi A . Autophagy as cell death and tumor suppressor mechanism. Oncogene 2004; 23: 2891–2906.
Abeliovich H, Dunn Jr WA, Kim J, Klionsky DJ . Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol 2000; 151: 1025–1034.
Tassa A, Roux MP, Attaix D, Bechet DM . Class III phosphoinositide 3-kinase–Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J 2003; 376: 577–586.
Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P . Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000; 275: 992–998.
Tsujimoto Y, Shimizu S . Another way to die: autophagic programmed cell death. Cell Death Differ 2005; 12 (Suppl 2): 1528–1534.
Vachon PH, Harnois C, Grenier A, Dufour G, Bouchard V, Han J et al. Differentiation state-selective roles of p38 isoforms in human intestinal epithelial cell anoikis. Gastroenterology 2002; 123: 1980–1991.
Vom Dahl S, Dombrowski F, Schmitt M, Schliess F, Pfeifer U, Ussinger D . Cell hydration controls autophagosome formation in rat liver in a microtubule-dependent way downstream from p38MAPK activation. Biochem J 2001; 354: 31–36.
Prick T, Thumm M, Kohrer K, Haussinger D, Vom Dahl S . In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem J 2006; 394: 153–161.
Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T . LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117: 2805–2812.
Codogno P, Meijer AJ . Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 2005; 12 (Suppl 2): 1509–1518.
Marx J . Autophagy: is it cancer's friend or foe? Science 2006; 312: 1160–1161.
Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res 2005; 65: 3336–3346.
Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S . Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ 2004; 11: 448–457.
Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I . Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res 2003; 63: 2103–2108.
Katayama M, Kawaguchi T, Berger MS, Pieper RO . DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ 2006; 1 September; (E-pub ahead of print).
Herman-Antosiewicz A, Johnson DE, Singh SV . Sulforaphane causes autophagy to inhibit release of cytochrome c and apoptosis in human prostate cancer cells. Cancer Res 2006; 66: 5828–5835.
Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A . Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 2006; 22 September; (E-pub ahead of print).
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64.
Ten Hove T, van den Blink B, Pronk I, Drillenburg P, Peppelenbosch MP, van Deventer SJ . Dichotomal role of inhibition of p38 MAPK with SB 203580 in experimental colitis. Gut 2002; 50: 507–512.
Prakash J, Sandovici M, Saluja V, Lacombe M, Schaapveld RQ, de Borst M et al. Intracellular delivery of the P38 MAPK inhibitor SB202190 in renal tubular cells: a novel strategy to treat renal fibrosis. J Pharmacol Exp Ther 2006; 319: 8–19.
Corin I, Di Giacomo MC, Lastella P, Bagnulo R, Guanti G, Simone C . Tumor-specific hyperactive low-molecular-weight cyclin E isoforms detection and characterization in non-metastatic colorectal tumors. Cancer Biol Ther 2006; 5: 198–203.
Acknowledgements
We thank Marie Basso for her editorial assistance; Drs FP Jori, N Resta and A Stella, for their helpful discussion during the preparation of the manuscript. This work was partly supported by a grant from the Associazione Italiana Ricerca sul Cancro (to AM) and a grant from Fondazione CARIME (to GG).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by Y Tsujimoto
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Comes, F., Matrone, A., Lastella, P. et al. A novel cell type-specific role of p38α in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ 14, 693–702 (2007). https://doi.org/10.1038/sj.cdd.4402076
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402076
Keywords
This article is cited by
-
Unexpected inhibition of the lipid kinase PIKfyve reveals an epistatic role for p38 MAPKs in endolysosomal fission and volume control
Cell Death & Disease (2024)
-
Uncoupling p38α nuclear and cytoplasmic functions and identification of two p38α phosphorylation sites on β-catenin: implications for the Wnt signaling pathway in CRC models
Cell & Bioscience (2023)
-
A new perspective on the autophagic and non-autophagic functions of the GABARAP protein family: a potential therapeutic target for human diseases
Molecular and Cellular Biochemistry (2023)
-
Role of p38 MAP kinase in cancer stem cells and metastasis
Oncogene (2022)
-
Pharmacological targeting of the novel β-catenin chromatin-associated kinase p38α in colorectal cancer stem cell tumorspheres and organoids
Cell Death & Disease (2021)