Visual Overview
Abstract
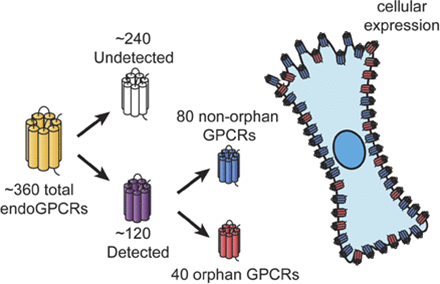
G protein–coupled receptors (GPCRs), the largest family of signaling receptors in the human genome, are also the largest class of targets of approved drugs. Are the optimal GPCRs (in terms of efficacy and safety) currently targeted therapeutically? Especially given the large number (∼120) of orphan GPCRs (which lack known physiologic agonists), it is likely that previously unrecognized GPCRs, especially orphan receptors, regulate cell function and can be therapeutic targets. Knowledge is limited regarding the diversity and identity of GPCRs that are activated by endogenous ligands and that native cells express. Here, we review approaches to define GPCR expression in tissues and cells and results from studies using these approaches. We identify problems with the available data and suggest future ways to identify and validate the physiologic and therapeutic roles of previously unrecognized GPCRs. We propose that a particularly useful approach to identify functionally important GPCRs with therapeutic potential will be to focus on receptors that show selective increases in expression in diseased cells from patients and experimental animals.
Footnotes
- Received January 23, 2015.
- Accepted March 2, 2015.
Work in the authors’ laboratory on this topic has been supported by research and training grants from the National Institutes of Health [Grants CA189477, CA121938, GM68524, HL091061, HL066941, HL007444, and GM-68524], the Department of Defense [Grant W81XWH-14-1-0372], and with financial assistance from Roche, Pfizer-CovX, Bristol Myers Squibb, the American Heart Association, and an ASPET-Astellas Award in Translational Pharmacology.
- Copyright © 2015 by The American Society for Pharmacology and Experimental Therapeutics
MolPharm articles become freely available 12 months after publication, and remain freely available for 5 years.Non-open access articles that fall outside this five year window are available only to institutional subscribers and current ASPET members, or through the article purchase feature at the bottom of the page.
|