Abstract
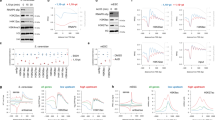
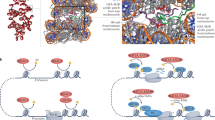
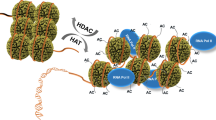
The amino termini of histones extend from the nucleosomal core and are modified by acetyltransferases and deacetylases during the cell cycle. These acetylation patterns may direct histone assembly and help regulate the unfolding and activity of genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Allfrey, V. G. in Chromatin and Chromosome Structure (eds Li, H. J. & Eckhardt, R.) 167–191 (Academic, New York, (1977)).
Hebbes, T. R., Thorne, A. W. & Crane-Robinsson, C. Adirect link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1402 (1988).
McGhee, J. D. & Felsenfeld, G. Nucleosome structure. Annu. Rev. Biochem. 49, 1115–1156 (1980).
Gross, D. A. & Garrard, W. T. Nucleosome hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197 (1988).
Norton, V. G., Marvin, K. W., Yau, P. & Bradbury, E. M. Nucleosome linking number change controlled by acetylation of histones H3 and H4 J. Biol. Chem. 265, 19848–19852 (1990).
Lee, D. Y., Hayes, J. J., Pruss, D. & Wolffe, A. P. Apositive role for histone acetylation in transcription factor access to DNA. Cell 72, 73–84 (1993).
Clarke, D. J., O'Neill, L. P. & Turner, B. M. Selective use of H4 acetylation sites in the yeast Saccharomyces cerevisiae. Biochem. J. 294, 557–561 (1993).
O'Neill, L. P. & Turner, B. M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14, 3946–3957 (1995).
Jeppesen, P. & Turner, B. M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74, 281–289 (1993).
Turner, B. M. et al. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69, 375–384 (1992).
Braunstein, M. et al. Efficient transcriptional silencing in Saccharomyces cerevisiae requires heterochromatin histone acetylation pattern. Mol. Cell Biol. 16, 4349–4356 (1996).
Sobel, R. E. et al. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA 92, 1237–1241 (1995).
Kuo, M. et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 (1966).
Kaufman, P. D. et al. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81, 1105–1114 (1995).
Verreault, A., Kaufman, P. D., Kobayashi, R. & Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 (1996).
Kayne, P. S. et al. Extremely conserved histone N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55, 27–39 (1988).
Megee, P. C., Morgan, B. A., Mittman, B. A. & Smith, M. M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247, 841–845 (1990).
Ling, X. et al. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 10, 686–699 (1996).
Freeman, L., Kurumizaka, H. & Wolffe, A. P. Functional domains for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proc. Natl Acad. Sci. USA 93, 12780–12785 (1996).
Ma, X., Wu, J., Altheim, B., Schultz, M. C. & Grunstein, M. Histone H4 acetylation sites that are required for deposition in vivo. Proc. Natl Acad. Sci. USA(submitted).
Ishimi, Y. & Kikuchi, A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266, 7025–7029 (1991).
Ito, T. et al. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16, 3112–3124 (1996).
Kim, U.-J., Han, M., Kayne, P. & Grunstein, M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7, 2211–2219 (1988).
Kaufman, P. D., Kobayashi, R. & Stillman, B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in S. cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11, 345–357 (1997).
Enomoto, S. et al. RLF1, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11, 358–370 (1997).
Kleff, S., Andrulis, E. D., Anderson, C. W. & Sternglanz, R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270, 24674–24677 (1997).
Parthun, M. R., Widom, J. & Gottschling, D. E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 (1996).
Taunton, J., Hassig, C. A. & Schreiber, S. L. Amammalian histone deacetylase related to the yeast transcriptional regulator RPD3p. Science 272, 408–411 (1996).
Tyler, J. K. et al. The p55 unit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 16, 6149–6159 (1996).
Johnson, L. M., Kayne, P. S., Kahn, E. S. & Grunstein, M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 87, 6286–6290 (1990).
Thompson, J. S., Ling, X. & Grunstein, M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369, 245–247 (1994).
Strahl-Bolsinger, S., Hecht, A., Luo, K. & Grunstein, M. SIR2 and SIR4 interactions differ in more and extended telomeric heterochromatin of yeast. Genes Dev. 11, 83–93 (1997).
Thompson, J. S., Hecht, A. & Grunstein, M. Histones and the regulation of heterochromatin in yeast. Cold Spring Harb. Symp. Quant. Biol. 53, 247–256 (1993).
Rundlett, S. E. et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA 93, 14503–14508 (1996).
Vannier, D., Balderes, D. & Shore, D. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in Saccharomyces cerevisiae. Genetics 144, 1343–1353 (1996).
De Rubertis, F. et al. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384, 589–591 (1996).
Vidal, M. & Gaber, R. F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 6317–6327 (1991).
Van Lint, C., Emiliani, S. & Verdin, E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Exp. 5, 245–253 (1996).
Locke, J., Kotarski, M. A. & Tartof, K. D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effects. Genetics 120, 181–198 (1988).
Renauld, H. et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7, 1133–1145 (1993).
Hecht, A., Strahl-Bolsinger, S. & Grunstein, M. Spreading of transcriptional repression by SIR from telomeric heterochromatin. Nature 383, 92–96 (1996).
Wilson, C. J. et al. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84, 235–244 (1996).
Grunstein, M. Histone function in transcription. Annu. Rev. Cell Biol. 6, 643–678 (1990).
Chasman, D. I. et al. Ayeast protein that influences the chromatin structure of UASg and functions as a powerful auxiliary gene activator. Genes Dev. 4, 503–514 (1990).
Roth, S. Y., Shimizu, M., Johnson, L., Grunstein, M. & Simpson, R. T. Stable nucleosome positioning and complete repression by the yeast α2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 6, 411–425 (1992).
Fisher-Adams, G. & Grunstein, M. Yeast histone H4 and H3 N termini have different effects on the chromatin structure of the GAL1 promoter. EMBO J. 14, 1468–1477 (1995).
Durrin, L. K., Mann, R. K., Kayne, P. S. & Grunstein, M. Yeast histone H4 N terminal sequence is required for promoter activation in vivo. Cell 65, 1023–1031 (1991).
Wan, J. S., Mann, R. K. & Grunstein, M. Yeast histone H3 and H4 N termini function through different GAL1 regulatory elements to repress and activate transcription. Proc. Natl Acad. Sci. USA 92, 5664–5668 ((1995)).
Brownell, J. E. et al. Tetrahymena histone acetyltransferase A: a homologue to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (1996).
Horiuchi, J., Silverman, N., Marcus, G. A. & Guarente, L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA3 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15, 1203–1209 (1995).
Georgakopoulos, T., Gounalaki, N. & Thireos, G. Genetic evidence for the interaction of yeast transcriptional co-activator proteins GCN5 and ADA2. Mol. Gen. Genet. 246, 723–728 (1995).
Candau, R., Zhou, J.-X., Allis, C. D. & Berger, S. L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16, 555–565 (1997).
Chakravarti, D. et al. Role of CBP/P300 in nuclear receptor signalling. Nature 383, 99–103 (1996).
Yang, X., Ogryzko, V. V., Nishikawa, J., Howard, B. & Nakatani, Y. Ap300/CBO-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324 (1996).
Ogryzko, V. V., Shiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996).
Bannister, A. J. & Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 (1996).
Borrelli, E., Hen, R. & Chambon, P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature 312, 608–612 (1984).
Ptashne, M. & Gann, A. Transcriptional activation by recruitment. Nature 386, 569–576 (1997).
Mizzen, C. A. et al. The TAF(II) 250 subunit of TFIID has histone acetyltransferase activity. Cell 87, 1261–1270 (1996).
Gaudreau, L., Schmid, A., Blaschke, D., Ptashne, M. & Horz, W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89, 55–62 (1997).
Yang, W. M., Inouye, C., Zeng, Y., Bearss, D. & Seto, E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA 93, 12845–12850 (1996).
Carmen, A. A., Rundlett, S. E. & Grunstein, M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271, 15837–15844 (1996).
Stillman, D. J., Dorland, S. & Yu, Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics 136, 781–788 (1994).
Ayer, D. E. et al. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80, 767–776 (1996).
Schreiber-Agus, N. et al. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor Sin3. Cell 80, 777–786 (1995).
Wolffe, A. P. Histone-deacetylase: A regulator of transcription. Science 272, 371–372 (1996).
Hassig, C. A., Fleischer, T. C., Billin, A. N., Schreiber, S. L. & Ayer, D. E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 83, 341–347 (1997).
Laherty, C. D. et al. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 83, 349–356 (1997).
Heinzel, T. et al. Acomplex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48 (1997).
Alland, L. et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55 (1997).
Nagy, L. et al. Nuclear receptor repression mediated by a complex containing SMRT, mSIN3A and histone deacetylase. Cell 83, 373–380 (1997).
Kadosh, D. & Struhl, K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd4 histone deacetylase to target promoters. Cell 83, 365–371 (1997).
Zhang, Y., Iratni, R., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 83, 357–364 (1997).
Pazin, M. J. & Kadonaga, J. T. What's up and down with histone deacetylation and transcription? Cell 89, 325–328 (1997).
Darkin-Rattray, S. J. et al. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl Acad. Sci. USA 93, 13143–13147 (1996).
Acknowledgements
I thank A. Berk, J. Kadonaga, G. Felsenfeld, S. Rundlett and A. Carmen for comments.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997). https://doi.org/10.1038/38664
Issue Date:
DOI: https://doi.org/10.1038/38664
This article is cited by
-
The histone deacetylase UvHOS2 regulates vegetative growth, conidiation, ustilaginoidin synthesis, and pathogenicity in Ustilaginoidea virens
Phytopathology Research (2024)
-
Guiding the HBO1 complex function through the JADE subunit
Nature Structural & Molecular Biology (2024)
-
Epigenetics as a versatile regulator of fibrosis
Journal of Translational Medicine (2023)
-
Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
How protons pave the way to aggressive cancers
Nature Reviews Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.