Abstract
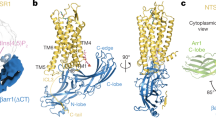
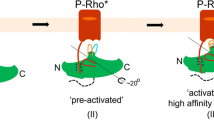
Arrestins interact with G-protein-coupled receptors (GPCRs) to block interaction with G proteins1,2 and initiate G-protein-independent signalling3. Arrestins have a bi-lobed structure that is stabilized by a long carboxy-terminal tail (C-tail), and displacement of the C-tail by receptor-attached phosphates activates arrestins for binding active GPCRs4. Structures of the inactive state of arrestin are available5,6, but it is not known how C-tail displacement activates arrestin for receptor coupling. Here we present a 3.0 Å crystal structure of the bovine arrestin-1 splice variant p44, in which the activation step is mimicked by C-tail truncation. The structure of this pre-activated arrestin is profoundly different from the basal state and gives insight into the activation mechanism. p44 displays breakage of the central polar core and other interlobe hydrogen-bond networks, leading to a ∼21° rotation of the two lobes as compared to basal arrestin-1. Rearrangements in key receptor-binding loops in the central crest region include the finger loop7,8,9, loop 139 (refs 8, 10, 11) and the sequence Asp 296–Asn 305 (or gate loop), here identified as controlling the polar core. We verified the role of these conformational alterations in arrestin activation and receptor binding by site-directed fluorescence spectroscopy. The data indicate a mechanism for arrestin activation in which C-tail displacement releases critical central-crest loops from restricted to extended receptor-interacting conformations. In parallel, increased flexibility between the two lobes facilitates a proper fitting of arrestin to the active receptor surface. Our results provide a snapshot of an arrestin ready to bind the active receptor, and give an insight into the role of naturally occurring truncated arrestins in the visual system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wilden, U., Hall, S. W. & Kuhn, H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl Acad. Sci. USA 83, 1174–1178 (1986)
Lohse, M. J., Benovic, J. L., Codina, J., Caron, M. G. & Lefkowitz, R. J. β-Arrestin: a protein that regulates β-adrenergic receptor function. Science 248, 1547–1550 (1990)
Shukla, A. K., Xiao, K. & Lefkowitz, R. J. Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci. 36, 457–469 (2011)
Gurevich, V. V., Hanson, S. M., Song, X., Vishnivetskiy, S. A. & Gurevich, E. V. The functional cycle of visual arrestins in photoreceptor cells. Prog. Retin. Eye Res. 30, 405–430 (2011)
Granzin, J. et al. X-ray crystal structure of arrestin from bovine rod outer segments. Nature 391, 918–921 (1998)
Hirsch, J. A., Schubert, C., Gurevich, V. V. & Sigler, P. B. The 2.8 Å crystal structure of visual arrestin: a model for arrestin’s regulation. Cell 97, 257–269 (1999)
Feuerstein, S. E. et al. Helix formation in arrestin accompanies recognition of photoactivated rhodopsin. Biochemistry 48, 10733–10742 (2009)
Hanson, S. M. et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc. Natl Acad. Sci. USA 103, 4900–4905 (2006)
Sommer, M. E., Farrens, D. L., McDowell, J. H., Weber, L. A. & Smith, W. C. Dynamics of arrestin-rhodopsin interactions: loop movement is involved in arrestin activation and receptor binding. J. Biol. Chem. 282, 25560–25568 (2007)
Kim, M. et al. Conformation of receptor-bound visual arrestin. Proc. Natl Acad. Sci. USA 109, 18407–18412 (2012)
Vishnivetskiy, S. A., Baameur, F., Findley, K. R. & Gurevich, V. V. Critical role of central 139-loop in stability and binding selectivity of arrestin-1. J. Biol. Chem.. http://dx.doi.org/10.1074/jbc.M113.450031 jbc M113.4 50031 (2013)
Gurevich, V. V. & Benovic, J. L. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 268, 11628–11638 (1993)
Schröder, K., Pulvermüller, A. & Hofmann, K. P. Arrestin and its splice variant Arr1–370A (p44). Mechanism and biological role of their interaction with rhodopsin. J. Biol. Chem. 277, 43987–43996 (2002)
Kirchberg, K. et al. Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc. Natl Acad. Sci. USA 108, 18690–18695 (2011)
Schleicher, A., Kühn, H. & Hofmann, K. P. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry 28, 1770–1775 (1989)
Smith, W. C. et al. A splice variant of arrestin. Molecular cloning and localization in bovine retina. J. Biol. Chem. 269, 15407–15410 (1994)
Pulvermüller, A. et al. Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry 36, 9253–9260 (1997)
Scheerer, P. et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008)
Choe, H. W. et al. Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011)
Gurevich, V. V. & Benovic, J. L. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J. Biol. Chem. 270, 6010–6016 (1995)
Vishnivetskiy, S. A. et al. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J. Biol. Chem. 275, 41049–41057 (2000)
Hanson, S. M. & Gurevich, V. V. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J. Biol. Chem. 281, 3458–3462 (2006)
Granzin, J. et al. Crystal structure of p44, a constitutively active splice variant of visual arrestin. J. Mol. Biol. 416, 611–618 (2012)
Mansoor, S. E., Dewitt, M. A. & Farrens, D. L. Distance mapping in proteins using fluorescence spectroscopy: the tryptophan-induced quenching (TrIQ) method. Biochemistry 49, 9722–9731 (2010)
Sommer, M. E., Hofmann, K. P. & Heck, M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nature Commun. 3, 995 (2012)
Sommer, M. E., Hofmann, K. P. & Heck, M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J. Biol. Chem. 286, 7359–7369 (2011)
Zhuang, T. et al. Involvement of distinct arrestin-1 elements in binding to different functional forms of rhodopsin. Proc. Natl Acad. Sci. USA 110, 942–947 (2012)
Gurevich, V. V. et al. Arrestin interactions with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, β2-adrenergic, and m2 muscarinic cholinergic receptors. J. Biol. Chem. 270, 720–731 (1995)
Azarian, S. M., King, A. J., Hallett, M. A. & Williams, D. S. Selective proteolysis of arrestin by calpain. Molecular characteristics and its effect on rhodopsin dephosphorylation. J. Biol. Chem. 270, 24375–24384 (1995)
Sommer, M. E., Smith, W. C. & Farrens, D. L. Dynamics of arrestin-rhodopsin interactions: acidic phospholipids enable binding of arrestin to purified rhodopsin in detergent. J. Biol. Chem. 281, 9407–9417 (2006)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Winn, M. D., Isupov, M. N. & Murshudov, G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Vaguine, A. A., Richelle, J. & Wodak, S. J. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D 55, 191–205 (1999)
Laskowski, R. A., Moss, D. S. & Thornton, J. M. Procheck: a program to check the stereo chemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. Errors in protein structures. Nature 381, 272 (1996)
McDonald, I. K. & Thornton, J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994)
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 (1995)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Poornam, G. P., Matsumoto, A., Ishida, H. & Hayward, S. A method for the analysis of domain movements in large biomolecular complexes. Proteins 76, 201–212 (2009)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002)
Sommer, M. E., Smith, W. C. & Farrens, D. L. Dynamics of arrestin-rhodopsin interactions: arrestin and retinal release are directly linked events. J. Biol. Chem. 280, 6861–6871 (2005)
Acknowledgements
We thank J. H. Park for help at the early stage of the project and B. Bauer, J. Engelmann, C. Koch and H. Seibel for technical assistance. We are grateful to U. Müller, M. Weiss and the scientific staff of the BESSY-MX/Helmholtz Zentrum Berlin für Materialien und Energie at beamlines BL 14.1, BL 14.2 and BL 14.3 operated by the Joint Berlin MX-Laboratory at the BESSY II electron storage ring (Berlin-Adlershof, Germany) and the scientific staff of the European Synchrotron Radiation Facility (ESRF, Grenoble) at beamlines ID14-1, ID 23-1, ID 23-2, ID 29S, ID 29 and ID 14-4 for continuous support. The data presented here were recorded at beamline ID 14-4 (ESRF, Grenoble). This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB449 to O.P.E., SFB740 to K.P.H. and O.P.E., SFB1078-B6 to P.S., SO1037/1-2 to M.E.S.), DFG Cluster of Excellence ‘Unifying Concepts in Catalysis’ (Research Field D3/E3-1 to P.S.), European Research Council (Advanced Investigator Grant (ERC-2009/249910-TUDOR to K.P.H.)), the Canada Excellence Research Chair program (to O.P.E.) and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012R1A1A2044752 to H.-W.C.). O.P.E. holds The Anne and Max Tanenbaum Chair in Neuroscience at the University of Toronto.
Author information
Authors and Affiliations
Contributions
K.P.H., O.P.E. and H.-W.C. designed the structural studies of p44. Y.J.K. performed p44 preparation, functional analysis and crystallization; Y.J.K., P.S. and H.-W.C. performed data collection and structural analysis; M.E.S. designed and performed functional assays and fluorescence measurements of labelled arrestin mutants; Y.J.K., K.P.H., P.S., H.-W.C. and M.E.S. analysed and interpreted data; M.E.S. wrote the paper with contributions from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-13, Supplementary Methods, Supplementary Tables 1-2, a Supplementary Discussion and Supplementary References. (PDF 7446 kb)
Rights and permissions
About this article
Cite this article
Kim, Y., Hofmann, K., Ernst, O. et al. Crystal structure of pre-activated arrestin p44. Nature 497, 142–146 (2013). https://doi.org/10.1038/nature12133
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12133
This article is cited by
-
Distinct activation mechanisms of β-arrestin-1 revealed by 19F NMR spectroscopy
Nature Communications (2023)
-
Agonist dependency of the second phase access of β-arrestin 2 to the heteromeric µ-V1b receptor
Scientific Reports (2021)
-
Structural aspects of rod opsin and their implication in genetic diseases
Pflügers Archiv - European Journal of Physiology (2021)
-
Phosphorylated peptide of G protein-coupled receptor induces dimerization in activated arrestin
Scientific Reports (2020)
-
Many faces of the GPCR-arrestin interaction
Archives of Pharmacal Research (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.