Key Points
-
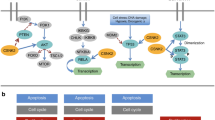
Processes that are relevant to cancer biology and that are regulated by 14-3-3 protein interactions include cell-cycle progression, apoptosis and mitogenic signalling.
-
14-3-3 proteins bind to protein ligands that have been phosphorylated on serine/threonine residues in a consensus binding motif. There are, however, a few proteins that associate with 14-3-3s independently of this motif.
-
14-3-3 proteins regulate other proteins by cytoplasmic sequestration, occupation of interaction domains and export or import sequences, prevention of degradation, activation/repression of enzymatic activity and transactivation, and facilitation of protein modifications. These effects are caused by 14-3-3-mediated conformational changes or steric hindrance.
-
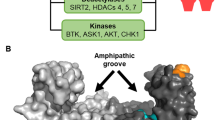
14-3-3 proteins form dimers that provide two binding sites for phosphoserine motifs in ligand proteins. They can therefore function as adaptor proteins, bringing two proteins that would not otherwise associate into close proximity. In addition, ligands with two 14-3-3-binding motifs might be bound with higher affinity by one 14-3-3 dimer.
-
In humans, seven expressed 14-3-3 isoforms have been identified, one of which — 14-3-3σ — is induced by DNA damage and is required for a stable G2 cell-cycle arrest in epithelial cells. The 14-3-3σ gene is directly regulated by p53; furthermore, 14-3-3σ is silenced by CpG methylation in a large proportion of carcinomas, which could be used for diagnosis.
-
14-3-3σ expression is restricted to epithelial cells and increases during epithelial differentiation. Inactivation of 14-3-3σ leads to immortalization of primary keratinocytes and prevents exit from the stem-cell compartment, indicating that this gene has tumour-suppressive properties.
-
Loss of 14-3-3σ expression sensitizes tumour cells to treatment with conventional cytostatic drugs. Modulation of 14-3-3σ activity might therefore be an attractive therapeutic approach.
Abstract
14-3-3 proteins regulate many cellular processes that are important in cancer biology, such as apoptosis and cell-cycle checkpoints. There are seven human 14-3-3 genes and one of these, 14-3-3σ, has been directly implicated in the aetiology of human cancer. Loss of 14-3-3σ expression sensitizes cancer cells to conventional anticancer agents, so its inhibition could be exploited for therapeutic purposes. Interference with 14-3-3 function as a therapeutic approach is being evaluated at present and, in the case of UCN-01, is under clinical investigation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Aitken, A. et al. 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17, 498–501 (1992).
Ferl, R. J., Manak, M. S. & Reyes, M. F. The 14-3–3s. Genome Biol. 3, reviews 3010.1–3010.7 (2002).
Wang, W. & Shakes, D. C. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43, 384–398 (1996).
Moore, B. E. & Perez, V. J. in Physiological and Biochemical Aspects of Nervous Integration (ed. Carlson, F. D.) 343–359 (Prentice–Hall, Englewood Cliffs, 1967).
Muslin, A. J., Tanner, J. W., Allen, P. M. & Shaw, A. S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 (1996). Shows the specificity of 14-3-3 proteins for ligands that contain an RSXpSXP-binding motif.
Yaffe, M. B. et al. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91, 961–971 (1997). Further analysis of the conditional 14-3-3-binding motif described in reference 5.
van Hemert, M. J., Steensma, H. Y. & van Heusden, G. P. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23, 936–946 (2001).
Aitken, A. et al. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 30, 351–360 (2002).
Fu, H., Subramanian, R. R. & Masters, S. C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 (2000).
Pawson, T., Gish, G. D. & Nash, P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11, 504–511 (2001).
Seimiya, H. et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19, 2652–2661 (2000).
Vincenz, C. & Dixit, V. M. 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J. Biol. Chem. 271, 20029–20034 (1996).
Xiao, B. et al. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376, 188–191 (1995). Together with reference 14, this paper describes the first crystal structures of the 14-3-3 dimer, which have important implications for 14-3–3 functions.
Liu, D. et al. Crystal structure of the ζ-isoform of the 14-3-3 protein. Nature 376, 191–194 (1995).
Obsil, T. et al. Crystal structure of the 14-3-3ζ:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 105, 257–267 (2001). Structural evidence for 14-3-3-induced conformational changes that regulate enzymatic activity.
Tzivion, G. & Avruch, J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277, 3061–3064 (2002).
Yaffe, M. B. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57 (2002).
Tzivion, G., Luo, Z. & Avruch, J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394, 88–92 (1998).
Dhillon, A. S. et al. A Raf-1 mutant that dissociates MEK/extracellular signal-regulated kinase activation from malignant transformation and differentiation but not proliferation. Mol. Cell. Biol. 23, 1983–1993 (2003).
Waterman, M. J., Stavridi, E. S., Waterman, J. L. & Halazonetis, T. D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nature Genet. 19, 175–178 (1998). The first evidence implicating 14-3-3 proteins in the regulation of p53 activity.
Stavridi, E. S., Chehab, N. H., Malikzay, A. & Halazonetis, T. D. Substitutions that compromise the ionizing radiation-induced association of p53 with 14-3-3 proteins also compromise the ability of p53 to induce cell cycle arrest. Cancer Res. 61, 7030–7033 (2001).
Zha, J. et al. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL . Cell 87, 619–628 (1996). Proposal of the 'sequestration model' to explain the anti-apoptotic activity of 14-3-3 proteins.
Lopez-Girona, A., Furnari, B., Mondesert, O. & Russell, P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397, 172–175 (1999).
Rittinger, K. et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153–166 (1999).
Yang, J., Winkler, K., Yoshida, M. & Kornbluth, S. Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 18, 2174–2183 (1999).
Zeng, Y. & Piwnica-Worms, H. DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding. Mol. Cell. Biol. 19, 7410–7419 (1999).
Kumagai, A. & Dunphy, W. G. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13, 1067–1072 (1999).
Brunet, A. et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156, 817–828 (2002). Together with references 25–27, this study showed that 14-3-3 proteins are 'molecular chauffeurs' rather than attachable nuclear export signals.
Muslin, A. J. & Xing, H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12, 703–709 (2000).
Grozinger, C. M. & Schreiber, S. L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA 97, 7835–7840 (2000).
Ellis, J. J. et al. CaM kinase IIδC phosphorylation of 14-3-3β in vascular smooth muscle cells: activation of class II HDAC repression. Mol. Cell. Biochem. 242, 153–161 (2003).
Prymakowska-Bosak, M. et al. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14. 3. 3 proteins. Mol. Cell. Biol. 22, 6809–6019 (2002).
Kanai, F. et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 (2000).
Cahill, C. M. et al. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 276, 13402–13410 (2001).
Fujita, N., Sato, S., Katayama, K. & Tsuruo, T. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 277, 28706–28713 (2002).
Peng, C. Y. et al. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277, 1501–1505 (1997). Together with reference 66, this paper describes a molecular mechanism for 14-3-3 protein function in the regulation of the mammalian cell cycle for the first time.
Liu, Y. C. et al. Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem. 272, 9979–9985 (1997).
Craparo, A., Freund, R. & Gustafson, T. A. 14-3-3ε interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 272, 11663–11669 (1997).
Forrest, A. & Gabrielli, B. Cdc25B activity is regulated by 14-3-3. Oncogene 20, 4393–4401 (2001).
Chen, M. S., Ryan, C. E. & Piwnica-Worms, H. chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 23, 7488–7497 (2003).
Braselmann, S. & McCormick, F. Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J. 14, 4839–4848 (1995).
Van Der Hoeven, P. C. et al. 14-3-3 isotypes facilitate coupling of protein kinase C-ζ to Raf-1: negative regulation by 14-3-3 phosphorylation. Biochem. J. 345, 297–306 (2000).
Dent, P. et al. Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science 268, 1902–1906 (1995).
Toyo-oka, K. et al. 14-3-3ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller–Dieker syndrome. Nature Genet. 34, 274–285 (2003).
Sehnke, P. C., DeLille, J. M. & Ferl, R. J. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14 (Suppl.), S339–S354 (2002).
Shen, Y. H. et al. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell 14, 4721–4733 (2003).
Woodcock, J. M. et al. The dimeric versus monomeric status of 14-3–3ζ is controlled by phosphorylation of Ser58 at the dimer interface. J. Biol. Chem. 278, 36323–36327 (2003).
Powell, D. W. et al. Proteomic identification of 14-3-3ζ as a mitogen-activated protein kinase-activated protein kinase 2 substrate: role in dimer formation and ligand binding. Mol. Cell. Biol. 23, 5376–5387 (2003).
Megidish, T. et al. A novel sphingosine-dependent protein kinase (SDK1) specifically phosphorylates certain isoforms of 14-3-3 protein. J. Biol. Chem. 273, 21834–21845 (1998).
Powell, D. W. et al. Identification of 14-3-3ζ as a protein kinase B/Akt substrate. J. Biol. Chem. 277, 21639–21642 (2002).
Urano, T. et al. Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature 417, 871–875 (2002). Demonstration of another level of regulation by 14-3-3σ, and presumably by 14-3-3 proteins in general: ubiquitylation-mediated proteasomal degradation initiated by the E3 ligase EFP.
Hermeking, H. et al. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3–11 (1997). The first demonstration of a direct connection between 14-3-3 genes and tumour suppression by p53.
Shiloh, Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer 3, 155–168 (2003).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Smits, V. A. & Medema, R. H. Checking out the G(2)/M transition. Biochim. Biophys. Acta 1519, 1–12 (2001).
Graves, P. R., Lovly, C. M., Uy, G. L. & Piwnica-Worms, H. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20, 1839–1851 (2001).
Peng, C. Y. et al. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 9, 197–208 (1998).
Bulavin, D. V. et al. Dual phosphorylation controls Cdc25 phosphatases and mitotic entry. Nature Cell Biol. 5, 545–551 (2003). CDC2-mediated phosphorylation of Ser214 on CDC25C inhibits 14-3-3 binding by blocking phosphorylation on Ser216.
Manke, I. A. & Yaffe, M. B. Chk'n out in mitosis. Cell Cycle 2, 236–237 (2003).
Bulavin, D. V. et al. Phosphorylation of Xenopus Cdc25C at Ser285 interferes with ability to activate a DNA damage replication checkpoint in pre-midblastula embryos. Cell Cycle 2 263–266 (2003).
Davezac, N. et al. Regulation of CDC25B phosphatases subcellular localization. Oncogene 19, 2179–2185 (2000).
Lee, J., Kumagai, A. & Dunphy, W. G. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol. Biol. Cell 12, 551–563 (2001).
Rothblum-Oviatt, C. J., Ryan, C. E. & Piwnica-Worms, H. 14-3-3 binding regulates catalytic activity of human Wee1 kinase. Cell Growth Differ. 12, 581–589 (2001).
Su, T. T. et al. Cell cycle roles for two 14-3-3 proteins during Drosophila development. J. Cell Sci. 114, 3445–3454 (2001).
Ford, J. C. et al. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265, 533–535 (1994). First demonstration of the involvement of 14-3-3 genes in cell-cycle regulation and checkpoints.
Sanchez, Y. et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277, 1497–1501 (1997).
Zhou, B. B. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000).
Bulavin, D. V. et al. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411, 102–107 (2001).
Jiang, K. et al. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser345. J. Biol. Chem. 278, 25207–25217 (2003).
Laronga, C., Yang, H. Y., Neal, C. & Lee, M. H. Association of the cyclin-dependent kinases and 14-3-3σ negatively regulates cell cycle progression. J. Biol. Chem. 275, 23106–23112 (2000).
Yang, H. Y. et al. 14-3-3σ positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 23, 7096–7107 (2003).
Yarden, R. I. et al. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nature Genet. 30, 285–289 (2002). 14-3-3σ is an integral part of BRCA1-induced G2 arrest.
Green, D. R. & Evan, G. I. A matter of life and death. Cancer Cell 1, 19–30 (2002).
Cory, S. & Adams, J. M. The Bcl2 family: regulators of the cellular life-or-death switch. Nature Rev. Cancer 2, 647–656 (2002).
Li, J. et al. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res. 58, 5667–5672 (1998).
Gross, A., McDonnell, J. M. & Korsmeyer, S. J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13, 1899–1911 (1999).
Datta, S. R. et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6, 41–51 (2000). 14-3-3 binding facilitates PKA phosphorylation of BAD, which leads to dissociation of BCL2.
Dudek, H. et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275, 661–665 (1997).
Yano, S., Tokumitsu, H. & Soderling, T. R. Calcium promotes cell survival through CaM-K kinase activation of the protein-kinase-B pathway. Nature 396, 584–587 (1998).
Sun, H. et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl Acad. Sci. USA 96, 6199–6204 (1999).
Wang, H. G. et al. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284, 339–343 (1999).
Chiang, C. W. et al. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin-3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood 97, 1289–1297 (2001).
Konishi, Y., Lehtinen, M., Donovan, N. & Bonni, A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol. Cell 9, 1005–1016 (2002). The first demonstration of the potential of regulated 14-3-3 association to integrate signals from different pathways.
Brunet, A. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999).
Basu, S. et al. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 (2003).
Nomura, M. et al. 14-3-3 interacts directly with and negatively regulates pro-apoptotic Bax. J. Biol. Chem. 278, 2058–2065 (2003).
Samuel, T. et al. The G2/M regulator 14-3-3σ prevents apoptosis through sequestration of Bax. J. Biol. Chem. 276, 45201–45206 (2001).
Zhu, J., Jiang, J., Zhou, W. & Chen, X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 58, 5061–5065 (1998).
Westfall, M. D., Mays, D. J., Sniezek, J. C. & Pietenpol, J. A. The ΔNp63α phosphoprotein binds the p21 and 14-3-3σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay–Wells syndrome-derived mutations. Mol. Cell. Biol. 23, 2264–2276 (2003).
Aprelikova, O. et al. BRCA1 is a selective co-activator of 14-3-3σ gene transcription in mouse embryonic stem cells. J. Biol. Chem. 276, 25647–25650 (2001).
Chan, T. A. et al. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 401, 616–620 (1999). The first knockout study of 14-3-3 function in mammalian cells.
Dhar, S. et al. Inactivation of 14-3-3σ influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol. Cell. Biol. 20, 7764–7772 (2000).
Pellegrini, G. et al. p63 identifies keratinocyte stem cells. Proc. Natl Acad. Sci. USA 98, 3156–3161 (2001).
Perez-Losada, J. & Balmain, A. Stem-cell hierarchy in skin cancer. Nature Rev. Cancer 3, 434–443 (2003).
Owens, D. M. & Watt, F. M. Contribution of stem cells and differentiated cells to epidermal tumours. Nature Rev. Cancer 3, 444–451 (2003).
Dellambra, E. et al. Downregulation of 14-3-3σ prevents clonal evolution and leads to immortalization of primary human keratinocytes. J. Cell Biol. 149, 1117–1130 (2000). 14-3-3σ antisense expression overrides senescence.
Ku, N. O., Liao, J. & Omary, M. B. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 17, 1892–1906 (1998).
Chan, T. A. et al. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 14, 1584–1588 (2000).
Takihara, Y., Matsuda, Y. & Hara, J. Role of the β-isoform of 14-3-3 proteins in cellular proliferation and oncogenic transformation. Carcinogenesis 21, 2073–2077 (2000).
Sugiyama, A. et al. Forced expression of antisense 14-3-3β RNA suppresses tumor cell growth in vitro and in vivo. Carcinogenesis 24, 1549–1559 (2003).
Martin, D., Brown-Luedi, M. & Chiquet-Ehrismann, R. Tenascin-C signaling through induction of 14-3-3τ. J. Cell Biol. 160, 171–175 (2003).
Nakanishi, K. et al. Elevated expression levels of the 14-3-3 family of proteins in lung cancer tissues. Hum. Antibodies 8, 189–194 (1997).
Villaret, D. B. et al. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope 110, 374–381 (2000).
Prasad, G. L., Valverius, E. M., McDuffie, E. & Cooper, H. L. Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 3, 507–513 (1992).
Ferguson, A. T. et al. High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc. Natl Acad. Sci. USA 97, 6049–6054 (2000). First demonstration of 14-3-3σ CpG methylation during tumour development.
Suzuki, H. et al. Inactivation of the 14-3-3σ gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 60, 4353–4357 (2000).
Iwata, N. et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3σ gene in human hepatocellular carcinoma. Oncogene 19, 5298–5302 (2000).
Osada, H. et al. Frequent and histological type-specific inactivation of 14-3-3σ in human lung cancers. Oncogene 21, 2418–2424 (2002).
Gasco, M. et al. Coincident inactivation of 14-3-3σ and p16INK4a is an early event in vulval squamous neoplasia. Oncogene 21, 1876–1881 (2002).
Gasco, M. et al. Epigenetic inactivation of 14-3-3σ in oral carcinoma: association with p16INK4a silencing and human papillomavirus negativity. Cancer Res. 62, 2072–2076 (2002).
Lodygin, D. et al. Analysis of 14-3-3σ expression in hyper-proliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene 22, 5519–5524 (2003). Shows that there is 14-3-3σ silencing in BCC, whereas other skin diseases associated with hyperproliferative keratinocytes retain 14-3-3σ expression.
Yatabe, Y. et al. Decreased expression of 14-3-3σ in neuroendocrine tumors is independent of origin and malignant potential. Oncogene 21, 8310–8319 (2002).
Umbricht, C. B. et al. Hypermethylation of 14-3-3σ (stratifin) is an early event in breast cancer. Oncogene 20, 3348–3353 (2001).
Tokugawa, T., Sugihara, H., Tani, T. & Hattori, T. Modes of silencing of p16 in development of esophageal squamous cell carcinoma. Cancer Res. 62, 4938–4944 (2002).
Ivanova, T. et al. Methylation and silencing of the retinoic acid receptor-β2 gene in cervical cancer. BMC Cancer 2, 4 (2002).
Li, Z. et al. Biallelic inactivation of the thyroid hormone receptor β1 gene in early stage breast cancer. Cancer Res. 62, 1939–1943 (2002).
Rhee, I. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556 (2002).
Bachman, K. E. et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 3, 89–95 (2003).
Rountree, M. R., Bachman, K. E., Herman, J. G. & Baylin, S. B. DNA methylation, chromatin inheritance, and cancer. Oncogene 20, 3156–3165 (2001).
Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 3, 415–428 (2002).
Ling, G. et al. PATCHED and p53 gene alterations in sporadic and hereditary basal cell cancer. Oncogene 20, 7770–7778 (2001).
Laird, P. W. The power and the promise of DNA methylation markers. Nature Rev. Cancer 3, 253–266 (2003).
Shawver, L. K., Slamon, D. & Ullrich, A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell 1, 117–123 (2002).
Hermeking, H. The Myc oncogene as a cancer drug target. Curr. Cancer Drug Targets 3, 163–175 (2003).
Druker, B. J. Perspectives on the development of a molecularly targeted agent. Cancer Cell 1, 31–36 (2002).
Lodygin, D., Menssen, A. & Hermeking, H. Induction of the Cdk inhibitor p21 by LY83583 inhibits tumor cell proliferation in a p53-independent manner. J. Clin. Invest. 110, 1717–1727 (2002).
Xing, H. et al. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 19, 349–358 (2000).
Masters, S. C. et al. Survival-promoting functions of 14-3-3 proteins. Biochem. Soc. Trans. 30, 360–365 (2002). Innovative design of a pharmacological inhibitor of 14-3-3–ligand interactions.
Wang, B. et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 (1999).
Wurtele, M., Jelich-Ottmann, C., Wittinghofer, A. & Oecking, C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 22, 987–994 (2003). Stabilization, rather than inhibition, of 14-3-3–ligand interactions as a therapeutic strategy.
Graves, P. R. et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275, 5600–5605 (2000). Together with reference 133, this paper describes the identification of the CHK1–CDC25C–14-3-3 pathway as an important target of the anticancer drug UCN-01, which could be used in combination with classical cancer therapeutics.
Zhao, B. et al. Structural basis for Chk1 inhibition by UCN-01. J. Biol. Chem. 277, 46609–46615 (2002).
Busby, E. C. et al. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60, 2108–2112 (2000).
Zhao, H., Watkins, J. L. & Piwnica-Worms, H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl Acad. Sci. USA 99, 14795–14800 (2002).
Bunch, R. T. & Eastman, A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin. Cancer Res. 2, 791–797 (1996).
Shao, R. G. et al. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 57, 4029–4035 (1997).
Jackson, J. R. et al. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 60, 566–572 (2000).
Wang, Q. et al. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl Cancer Inst. 88, 956–965 (1996).
Senderowicz, A. M. Novel direct and indirect cyclin-dependent kinase modulators for the prevention and treatment of human neoplasms. Cancer Chemother. Pharmacol. 52 (Suppl. 1), S61–S73 (2003).
Tzivion, G., Luo, Z. J. & Avruch, J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3–3 partners in vivo. J. Biol. Chem. 275, 29772–29778 (2000).
Leffers, H. et al. Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. A member of a protein family that has been involved in the protein kinase C signalling pathway. J. Mol. Biol. 231, 982–998 (1993).
Acknowledgements
The author thanks A. Menssen, F. Melchior, L. Hengst and A. Benzinger for helpful comments. Work in the author's laboratory is supported by the Max-Planck-Society, Deutsche Krebshilfe, Thyssen-Foundation, Rudolf-Bartling-Foundation, and Studienstiftung des Deutschen Volkes.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Glossary
- SAGE
-
(Serial analysis of gene expression). Quantification of global gene-expression patterns based on large-scale sequencing of short sequence tags derived from the 3′ ends of messenger RNAs; this technique has been used extensively to characterize cancer-cell transcriptomes.
- MITOTIC CATASTROPHE
-
Cells that enter mitosis prematurely in the presence of DNA damage undergo an aberrant mitosis that results in cell death.
- MAINTENANCE METHYLASE DNMT1
-
Required for maintenance of the inherited methylation pattern; this enzyme uses hemimethylated DNA as a substrate after DNA replication.
- DE NOVO METHYLASE DNMT3B
-
Required for the de novo methylation that occurs during early development and gametogenesis.
Rights and permissions
About this article
Cite this article
Hermeking, H. The 14-3-3 cancer connection. Nat Rev Cancer 3, 931–943 (2003). https://doi.org/10.1038/nrc1230
Issue Date:
DOI: https://doi.org/10.1038/nrc1230
This article is cited by
-
KSR2-14–3-3ζ complex serves as a biomarker and potential therapeutic target in sorafenib-resistant hepatocellular carcinoma
Biomarker Research (2022)
-
14-3-3 Proteins are Potential Regulators of Liquid–Liquid Phase Separation
Cell Biochemistry and Biophysics (2022)
-
Loss of 4.1N in epithelial ovarian cancer results in EMT and matrix-detached cell death resistance
Protein & Cell (2021)
-
From plant physiology to pharmacology: fusicoccin leaves the leaves
Planta (2019)
-
NOTCH1 regulates the viability of cholangiocarcinoma cells via 14-3-3 theta
Journal of Cell Communication and Signaling (2019)