Abstract
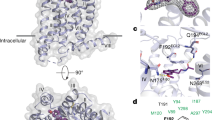
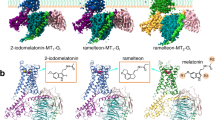
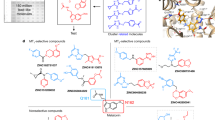
Melatonin (N-acetyl-5-methoxytryptamine) is a neurohormone that maintains circadian rhythms1 by synchronization to environmental cues and is involved in diverse physiological processes2 such as the regulation of blood pressure and core body temperature, oncogenesis, and immune function3. Melatonin is formed in the pineal gland in a light-regulated manner4 by enzymatic conversion from 5-hydroxytryptamine (5-HT or serotonin), and modulates sleep and wakefulness5 by activating two high-affinity G-protein-coupled receptors, type 1A (MT1) and type 1B (MT2)3,6. Shift work, travel, and ubiquitous artificial lighting can disrupt natural circadian rhythms; as a result, sleep disorders affect a substantial population in modern society and pose a considerable economic burden7. Over-the-counter melatonin is widely used to alleviate jet lag and as a safer alternative to benzodiazepines and other sleeping aids8,9, and is one of the most popular supplements in the United States10. Here, we present high-resolution room-temperature X-ray free electron laser (XFEL) structures of MT1 in complex with four agonists: the insomnia drug ramelteon11, two melatonin analogues, and the mixed melatonin–serotonin antidepressant agomelatine12,13. The structure of MT2 is described in an accompanying paper14. Although the MT1 and 5-HT receptors have similar endogenous ligands, and agomelatine acts on both receptors, the receptors differ markedly in the structure and composition of their ligand pockets; in MT1, access to the ligand pocket is tightly sealed from solvent by extracellular loop 2, leaving only a narrow channel between transmembrane helices IV and V that connects it to the lipid bilayer. The binding site is extremely compact, and ligands interact with MT1 mainly by strong aromatic stacking with Phe179 and auxiliary hydrogen bonds with Asn162 and Gln181. Our structures provide an unexpected example of atypical ligand entry for a non-lipid receptor, lay the molecular foundation of ligand recognition by melatonin receptors, and will facilitate the design of future tool compounds and therapeutic agents, while their comparison to 5-HT receptors yields insights into the evolution and polypharmacology of G-protein-coupled receptors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
03 May 2019
Change history: In this Letter, the rotation signs around 90°, 135° and 15° were missing and in the HTML, Extended Data Tables 2 and 3 were the wrong tables; these errors have been corrected online.
References
Pévet, P. Melatonin receptors as therapeutic targets in the suprachiasmatic nucleus. Expert Opin. Ther. Targets 20, 1209–1218 (2016).
Hardeland, R., Pandi-Perumal, S. R. & Cardinali, D. P. Melatonin. Int. J. Biochem. Cell Biol. 38, 313–316 (2006).
Dubocovich, M. L. & Markowska, M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine 27, 101–110 (2005).
Ganguly, S., Coon, S. L. & Klein, D. C. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 309, 127–137 (2002).
Tosini, G., Owino, S., Guillaume, J. L. & Jockers, R. Understanding melatonin receptor pharmacology: latest insights from mouse models, and their relevance to human disease. BioEssays 36, 778–787 (2014).
Dubocovich, M. L. et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol. Rev. 62, 343–380 (2010).
Stoller, M. K. Economic effects of insomnia. Clin. Ther. 16, 873–897, discussion 854 (1994).
Jockers, R. et al. Update on melatonin receptors: IUPHAR Review 20. Br. J. Pharmacol. 173, 2702–2725 (2016).
Zlotos, D. P. Recent progress in the development of agonists and antagonists for melatonin receptors. Curr. Med. Chem. 19, 3532–3549 (2012).
Clarke, T. C., Black, L. I., Stussman, B. J., Barnes, P. M. & Nahin, R. L. Trends in the use of complementary health approaches among adults: United States, 2002–2012. Natl. Health Stat. Report 79, 1–16 (2015).
Owen, R. T. Ramelteon: profile of a new sleep-promoting medication. Drugs Today (Barc) 42, 255–263 (2006).
Millan, M. J. et al. The novel melatonin agonist agomelatine (S20098) is an antagonist at 5-hydroxytryptamine2C receptors, blockade of which enhances the activity of frontocortical dopaminergic and adrenergic pathways. J. Pharmacol. Exp. Ther. 306, 954–964 (2003).
Guardiola-Lemaitre, B. et al. Agomelatine: mechanism of action and pharmacological profile in relation to antidepressant properties. Br. J. Pharmacol. 171, 3604–3619 (2014).
Johansson, L. C. et al. XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature https://doi.org/10.1038/s41586-019-1144-0 (2019).
Yin, J., Mobarec, J. C., Kolb, P. & Rosenbaum, D. M. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015).
Ballesteros, J. A. & Weinstein, H. in Methods in Neurosciences Vol. 25 (ed. Sealfon, S. C.) 366–428 (Academic, 1995).
Katritch, V. et al. Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014).
White, K. L. et al. Structural connection between activation microswitch and allosteric sodium site in GPCR signaling. Structure 26, 259–269.e5 (2018).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007).
Stauch, B. & Cherezov, V. Serial femtosecond crystallography of G protein-coupled receptors. Annu. Rev. Biophys. 47, 377–397 (2018).
Isberg, V. et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 44 (D1), D356–D364 (2016).
Bento, A. P. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–D1090 (2014).
Dubocovich, M. L. Luzindole (N-0774): a novel melatonin receptor antagonist. J. Pharmacol. Exp. Ther. 246, 902–910 (1988).
Reppert, S. M., Weaver, D. R., Ebisawa, T., Mahle, C. D. & Kolakowski, L. F. Jr Cloning of a melatonin-related receptor from human pituitary. FEBS Lett. 386, 219–224 (1996).
Clement, N. et al. Importance of the second extracellular loop for melatonin MT1 receptor function and absence of melatonin binding in GPR50. Br. J. Pharmacol. 175, 3281–3297 (2018).
Azmitia, E. C. Serotonin and brain: evolution, neuroplasticity, and homeostasis. Int. Rev. Neurobiol. 77, 31–56 (2007).
Tan, D. X. et al. Melatonin: a hormone, a tissue factor, an autocoid, a paracoid, and an antioxidant vitamin. J. Pineal Res. 34, 75–78 (2003).
Yu, H., Dickson, E. J., Jung, S. R., Koh, D. S. & Hille, B. High membrane permeability for melatonin. J. Gen. Physiol. 147, 63–76 (2016).
de la Fuente Revenga, M. et al. Novel N-acetyl bioisosteres of melatonin: melatonergic receptor pharmacology, physicochemical studies, and phenotypic assessment of their neurogenic potential. J. Med. Chem. 58, 4998–5014 (2015).
Peng, Y. et al. 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. Cell 172, 719–730.e14 (2018).
Pasqualetti, M. et al. Distribution and cellular localization of the serotonin type 2C receptor messenger RNA in human brain. Neuroscience 92, 601–611 (1999).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Johnson, M. et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 (2008).
Papadopoulos, J. S. & Agarwala, R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007).
Isberg, V. et al. Generic GPCR residue numbers — aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015).
Kullback, S. & Leibler, R. A. On information and sufficiency. Ann. Math. Stat. 22, 79–86 (1951).
White, J. F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012).
Shibata, Y. et al. Thermostabilization of the neurotensin receptor NTS1. J. Mol. Biol. 390, 262–277 (2009).
Roth, C. B., Hanson, M. A. & Stevens, R. C. Stabilization of the human β2-adrenergic receptor TM4–TM3–TM5 helix interface by mutagenesis of Glu1223.41, a critical residue in GPCR structure. J. Mol. Biol. 376, 1305–1319 (2008).
Bhattacharya, S., Hall, S. E. & Vaidehi, N. Agonist-induced conformational changes in bovine rhodopsin: insight into activation of G-protein-coupled receptors. J. Mol. Biol. 382, 539–555 (2008).
Klco, J. M., Nikiforovich, G. V. & Baranski, T. J. Genetic analysis of the first and third extracellular loops of the C5a receptor reveals an essential WXFG motif in the first loop. J. Biol. Chem. 281, 12010–12019 (2006).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protocols 4, 706–731 (2009).
Liu, W., Ishchenko, A. & Cherezov, V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protocols 9, 2123–2134 (2014).
Weierstall, U. et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309 (2014).
Boutet, S. & Williams, G. J. The Coherent X-ray Imaging (CXI) instrument at the Linac Coherent Light Source (LCLS). New J. Phys. 12, 035024 (2010).
Hart, P. et al. The Cornell-SLAC Pixel Array Detector at LCLS. SLAC-PUB https://ieeexplore.ieee.org/abstract/document/6551166 (2012).
Barty, A. et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 47, 1118–1131 (2014).
White, T. A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341 (2012).
Leslie, A. G. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 (2006).
Duisenberg, A. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Crystallogr. 25, 92–96 (1992).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
White, T. A. et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 49, 680–689 (2016).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
BUSTER v. 2.10.2 (Global Phasing Ltd., Cambridge, 2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Schüttelkopf, A. W. & van Aalten, D. M. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Alexandrov, A. I., Mileni, M., Chien, E. Y., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008).
Depreux, P. et al. Synthesis and structure-activity relationships of novel naphthalenic and bioisosteric related amidic derivatives as melatonin receptor ligands. J. Med. Chem. 37, 3231–3239 (1994).
Yous, S. et al. Novel naphthalenic ligands with high affinity for the melatonin receptor. J. Med. Chem. 35, 1484–1486 (1992).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Cock, P. J. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Henikoff, S. & Henikoff, J. G. Performance evaluation of amino acid substitution matrices. Proteins 17, 49–61 (1993).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Senes, A., Gerstein, M. & Engelman, D. M. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol. 296, 921–936 (2000).
O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. J. Cheminform. 3, 33 (2011).
Kozlikova, B. et al. CAVER Analyst 1.0: graphic tool for interactive visualization and analysis of tunnels and channels in protein structures. Bioinformatics 30, 2684–2685 (2014).
Schrödinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Abagyan, R., Totrov, M. & Kuznetsov, D. ICM—A new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 (1994).
Halgren, T. A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17, 490–519 (1996).
Zhang, H. et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544, 327–332 (2017).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Liu, W. et al. Serial femtosecond crystallography of G protein-coupled receptors. Science 342, 1521–1524 (2013).
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Acknowledgements
We thank M. Chu, C. Hanson, K. Villers, J. Velasquez, and H. Shaye for technical support, and D.R. Mende for useful discussion of sequence analysis. This research was supported by the National Institutes of Health (NIH) grants R35 GM127086 (V.C.), R21 DA042298 (W.L.), R01 GM124152 (W.L.), R01 MH112205 (B.L.R.), and U24DK116195 (B.L.R.), the NIMH Psychoactive Drug Screening Program contract (B.L.R.), F31-NS093917 (R.H.J.O.), the National Science Foundation (NSF) BioXFEL Science and Technology Center 1231306 (B.S., W.L., U.W., T.D.G., V.C.), EMBO ALTF 677-2014 (B.S.), HFSP long-term fellowship LT000046/2014-L (L.C.J.), and a postdoctoral fellowship from the Swedish Research Council (L.C.J.). C.G. thanks the SLAC National Accelerator Laboratory and the Department of Energy for financial support through the Panofsky fellowship. T.A.W. and W.B. acknowledge financial support from the Helmholtz Association via Programme-Oriented Funding. Parts of this research were carried out at the LCLS, a National User Facility operated by Stanford University on behalf of the US Department of Energy and supported by the US Department of Energy Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515.
Reviewer information
Nature thanks Christian Siebold, Ieva Sutkeviciute, Jean-Pierre Vilardaga and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
B.S., L.C.J., W.L. and V.C. conceived the project, analysed data and wrote the manuscript with contributions from all authors. B.S. and L.C.J. cloned and characterized the receptor, generated all constructs, crystallized the receptor, prepared all crystal samples and figures, solved and refined the structures, and assisted in generating mutant constructs for binding and functional analyses. B.S. designed thermostabilizing point mutations and performed sequence analysis. J.D.M., X.-P.H. and S.T.S. performed radioligand binding and functional experiments, assisted in generating mutant and wild-type constructs used for binding and functional analyses, and analysed all binding and functional data. A.I., N.M., A.S., L.Z. and W.L. assisted in XFEL sample preparation. G.W.H. performed structure refinement and quality control. B.S., L.C.J., A.B., L.Z., W.L. and V.C. collected XFEL data. C.G., W.B., T.A.W. and T.D.G. processed XFEL data and solved the indexing ambiguity. C.M. and U.W. operated the LCP injector during XFEL data collection. N.P. performed molecular docking and molecular dynamics calculations and assisted in preparing figures. J.M.G. assisted in docking calculations. V.K. supervised molecular docking and molecular dynamics calculations. R.H.J.O. assisted with molecular biology and generating mutant constructs. A.R.T. assisted with generating mutant constructs and functional experiments. S.Y. synthesized the bitopic compound, analysed data and edited the paper. R.C.S. contributed to study design and selection of chemical compounds for receptor stabilization and functional characterization, supervised protein expression and edited the paper. B.L.R. supervised pharmacological experiments and edited the paper. W.L. supervised the LCP crystallization and optimization experiments. V.C. coordinated and supervised the whole project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Crystals, ligand electron density maps, and packing of MT1.
a, b, Bright field (a) and cross-polarized (b) images of representative MT1–2-PMT crystals, optimized for synchrotron data collection (representing three independent crystallization setups). c, Cross-polarized image of representative MT1–ramelteon crystals used for XFEL data collection (representing two independent crystallization setups). d, 2mFo − DFc ligand electron density maps of MT1 co-crystallized with 2-PMT (orange), 2-iodomelatonin (yellow), and agomelatine (cyan), contoured at 1.0σ (grey mesh). e, 2mFo − DFc (blue, contoured at 1.0σ) and mFo − DFc (green/red, ±3.5σ) electron density maps of MT1–ramelteon (ligand purple, protein yellow) illustrating the small, unassigned electron density close to N2556.52 that is tentatively attributed to the essential additive 2-propan-ol. The distance from this electron density to the closest ligand atom is approximately 4.8 Å. f, Packing of MT1–PGS crystallized in the P4 21 2 space group. The receptor is shown in green and the PGS fusion protein is shown in purple. g, Simulated annealing mFo − DFc omit maps (green mesh) of 2-PMT (orange sticks), 2-iodomelatonin (yellow), and agomelatine (cyan), contoured at 3.0σ.
Extended Data Fig. 2 Molecular dynamics simulations.
a, b, Distance plots for interactions between residues in MT1 (N1624.60, atom type ND2 (Nδ); Q181ECL2, atom NE2 (Nε); N2556.52, atom ND2), and the closest oxygen atoms of the methoxy and acetyl groups, respectively, in the ligands melatonin (a) and 2-PMT (b) from three independent simulation runs. c, Distance histograms for interactions of methoxy with N1624.60 (left), and Q181ECL2 with the ligand acetyl tail (right), in melatonin and 2-PMT complexes. d, Hydration of residue N2556.52 over the course of a 1-µs simulation of the MT1–2-PMT complex from three independent simulations. e, Stability of ligand binding in simulations of MT1 complexes. Time dependence of r.m.s.d. for non-hydrogen atoms of melatonin shown for MT1–melatonin complex (left) and MT1–2-PMT complex (right). Three independent simulations of crystal construct (purple, blue, light blue) and crystal construct with N2556.52A mutation (orange, light orange, yellow) are shown, spanning 1.5 μs of cumulative time per system. Sampling rate was 10 frames per ns, and solid lines represent moving average values from 50 frames in all cases.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 and an Experimental Section describing synthesis of the bitopic ligand CTL 01-05-B-A05.
Rights and permissions
About this article
Cite this article
Stauch, B., Johansson, L.C., McCorvy, J.D. et al. Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288 (2019). https://doi.org/10.1038/s41586-019-1141-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1141-3
This article is cited by
-
Protective effects of melatonin on myocardial microvascular endothelial cell injury under hypertensive state by regulating Mst1
BMC Cardiovascular Disorders (2023)
-
Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding
Nature Communications (2023)
-
Fusion protein strategies for cryo-EM study of G protein-coupled receptors
Nature Communications (2022)
-
Serial femtosecond crystallography
Nature Reviews Methods Primers (2022)
-
Melatonin abolished proinflammatory factor expression and antagonized osteoarthritis progression in vivo
Cell Death & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.