-
PDF
- Split View
-
Views
-
Cite
Cite
Siyu Xia, Jing Feng, Lijun Lei, Jun Hu, Linjian Xia, Jun Wang, Yu Xiang, Lingjun Liu, Shan Zhong, Leng Han, Chunjiang He, Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes, Briefings in Bioinformatics, Volume 18, Issue 6, November 2017, Pages 984–992, https://doi.org/10.1093/bib/bbw081
Close - Share Icon Share
Abstract
Circular RNA (circRNA) is a group of RNA family generated by RNA circularization, which was discovered ubiquitously across different species and tissues. However, there is no global view of tissue specificity for circRNAs to date. Here we performed the comprehensive analysis to characterize the features of human and mouse tissue-specific (TS) circRNAs. We identified in total 302 853 TS circRNAs in the human and mouse genome, and showed that the brain has the highest abundance of TS circRNAs. We further confirmed the existence of circRNAs by reverse transcription polymerase chain reaction (RT-PCR). We also characterized the genomic location and conservation of these TS circRNAs and showed that the majority of TS circRNAs are generated from exonic regions. To further understand the potential functions of TS circRNAs, we identified microRNAs and RNA binding protein, which might bind to TS circRNAs. This process suggested their involvement in development and organ differentiation. Finally, we constructed an integrated database TSCD (Tissue-Specific CircRNA Database: http://gb.whu.edu.cn/TSCD) to deposit the features of TS circRNAs. This study is the first comprehensive view of TS circRNAs in human and mouse, which shed light on circRNA functions in organ development and disorders.
Introduction
Circular RNAs (circRNAs) are RNAs generated from back-splicing instead of the regular splicing [1, 2]. CircRNAs are prevalent in various species, from archaea, fungi to plants and animals. They are derived from diverse genomic regions, such as the intronic, exonic or intergenic region [3–7]. There are three types of biogenesis of circRNA. The first type is generated through the flanking intron sequence [8, 9]. The presence of reverse complementary sequences in introns might be paired and generate the ‘backspliced’ exons, and this noncanonical splicing depended on RNA-editing enzyme [9]. The second type is the formation of exon-containing lariat precursor, which is used more often in lower eukaryotes. This type of circRNAs do not have long introns [10]. The third type is companied by the interactions of RNA binding proteins (RBPs). Splicing factors (QKI and MBL) binding in both of the introns flank the exons that could interact and drive the circularization [8, 11].
Previous work revealed the functions of circRNAs mainly in three aspects. The first function is as miRNA sponge in transcription regulation [6, 12]. For example, circRNA Cdr1 works as a miR-7 sponge and regulates the insulin secretion and β cells renewal in diabetes [13]. While another miR-7 sponge circRNA ciRS-7 was reported highly expressed in human and mouse brain, which participates in regulating of neocortical and hippocampal neurons [14]. The interaction of mRNA, miRNA and circRNAs is also observed in liver cell lines, which respond to chemical carcinogenicity [15]. The second function is as a RBP sponge. For example, circRNA circ-Foxo3 can interact with anti-senescent protein ID-1, anti-stress proteins FAK and HIF1α and transcription factor E2F1, resulting in an increased cellular senescence [16]. CircRNA circ-Foxo3 can block the cell cycle progression by forming a complex with p21 and CDK2 [17]. The third function is related to transcription process. For example, the exon-intron circRNA ci-ankrd52 can bind to the RNA Pol II complex and acts as a cis-regulator of the transcription of the host gene ankrd52 [18].
High-throughput sequencing provided abundant resources for identification of circRNAs. Thousands of cell-specific circRNA loci have been discovered in human and mouse cell lines, suggesting that circRNAs may have cell-type or tissue specificity [19]. Furthermore, circRNA expressions are associated with spatial and temporal regulation, which was observed in fetal development by transcriptome sequencing, indicating the different spliceosome splice sites were used by developmentally induced circRNAs [20, 21]. Several circRNAs are identified to be negatively correlated to cancer cell proliferation [22]. Neural circRNAs have also been demonstrated to be involved in synapse formation and synaptic functions [23]. CircRNAs have also been used as a class of novel biomarkers for diagnosis of several diseases [24–27]. Despite the identification of large number of circRNAs, a comprehensive classification and characterization of circRNA in tissue and developmental process is still lacking.
Various algorithms have been developed to identify the circRNAs from RNA-seq data [6, 7, 28]. However, most of the algorithms focus on detecting the novel circRNA instead of tissue-specific (TS) circRNAs. There are few databases for circRNAs but either without the focus on TS features, such as circBase [29], or applied only one algorithm on polyA-enriched RNA-seq data in human, such as CircNet [30]. Here we identified the TS circRNAs in whole transcriptome level and characterized the features and functions of circRNAs across human adult and fetal tissues, as well as mouse tissues. We also constructed an integrated TS circRNA database TSCD (Tissue-Specific CircRNA Database; http://gb.whu.edu.cn/TSCD). Our results provided the information for the features and functions of TS circRNAs, thus benefiting the exploring of new RNA biomarkers in organ development and investigating the transcript process in organ differentiation and development disorders.
Materials and methods
Data collections of RNA-seq samples
We collected RNA-seq from ENCODE (https://www.encodeproject.org/) [31] and NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/). In total, we collected 16 adult human tissues (60 samples) and 10 fetal human tissues (28 samples) from ENCODE, 5 fetal human tissues (11 samples) from NCBI GEO data sets (GSE64283) and 9 mouse tissues from NCBI GEO data sets GSE61991 (15 samples) and GSE74747 (9 samples). All samples were prepared by total RNA with rRNA depletion, except samples from GSE74747, which were prepared without rRNA depletion. To be consistent with other samples, we used additional steps to remove the possible rRNAs in these samples by using SortMeRNA [32]. Detailed information for above 123 samples were listed in Table 1.
Tissue-specific circRNAs in the human and mouse genomes
| Species . | Tissue type . | Number of samples . | Number of TS circRNAs . | Average number of TS circRNAs per sample . |
|---|---|---|---|---|
| Human adult | Adipose | 8 | 11 020 | 1378 |
| Adrenal | 2 | 3594 | 1797 | |
| Blood vessel | 2 | 4464 | 2232 | |
| Esophagogastric | 4 | 8876 | 2219 | |
| Esophagus | 8 | 19 503 | 2438 | |
| Female gonad | 2 | 5455 | 2728 | |
| Heart | 5 | 15 278 | 3056 | |
| Intestine | 6 | 15 505 | 2584 | |
| Liver | 4 | 21 536 | 5384 | |
| Lung | 4 | 5230 | 1308 | |
| Mammary gland | 3 | 9193 | 3064 | |
| Pancreas | 2 | 1727 | 864 | |
| Skeletal muscle | 3 | 5546 | 1849 | |
| Skin | 2 | 3968 | 1984 | |
| Thyroid gland | 1 | 3599 | 3599 | |
| Tibial nerve | 4 | 6187 | 1547 | |
| Human fetus | Adrenal | 2 | 358 | 179 |
| Brain | 10 | 89 137 | 8914 | |
| Eye | 2 | 10 495 | 5248 | |
| Heart | 3 | 1608 | 536 | |
| Intestine | 2 | 390 | 195 | |
| Kidney | 2 | 319 | 160 | |
| Liver | 2 | 7906 | 3953 | |
| Lung | 2 | 559 | 280 | |
| Skeletal muscle | 2 | 12 932 | 6466 | |
| Skin | 2 | 9069 | 4535 | |
| Stomach | 2 | 7104 | 3552 | |
| Thyroid gland | 2 | 4190 | 2095 | |
| Tongue | 2 | 5024 | 2512 | |
| Umbilical cord | 2 | 1665 | 833 | |
| Uterus | 2 | 13 313 | 6657 | |
| Mouse | Brain | 4 | 5102 | 1276 |
| Heart | 4 | 797 | 199 | |
| Kidney | 1 | 624 | 624 | |
| Liver | 4 | 982 | 246 | |
| Lung | 4 | 1066 | 267 | |
| Skin | 1 | 806 | 806 | |
| Spleen | 1 | 546 | 546 | |
| Testis | 4 | 5374 | 1344 | |
| Thymus | 1 | 683 | 683 |
| Species . | Tissue type . | Number of samples . | Number of TS circRNAs . | Average number of TS circRNAs per sample . |
|---|---|---|---|---|
| Human adult | Adipose | 8 | 11 020 | 1378 |
| Adrenal | 2 | 3594 | 1797 | |
| Blood vessel | 2 | 4464 | 2232 | |
| Esophagogastric | 4 | 8876 | 2219 | |
| Esophagus | 8 | 19 503 | 2438 | |
| Female gonad | 2 | 5455 | 2728 | |
| Heart | 5 | 15 278 | 3056 | |
| Intestine | 6 | 15 505 | 2584 | |
| Liver | 4 | 21 536 | 5384 | |
| Lung | 4 | 5230 | 1308 | |
| Mammary gland | 3 | 9193 | 3064 | |
| Pancreas | 2 | 1727 | 864 | |
| Skeletal muscle | 3 | 5546 | 1849 | |
| Skin | 2 | 3968 | 1984 | |
| Thyroid gland | 1 | 3599 | 3599 | |
| Tibial nerve | 4 | 6187 | 1547 | |
| Human fetus | Adrenal | 2 | 358 | 179 |
| Brain | 10 | 89 137 | 8914 | |
| Eye | 2 | 10 495 | 5248 | |
| Heart | 3 | 1608 | 536 | |
| Intestine | 2 | 390 | 195 | |
| Kidney | 2 | 319 | 160 | |
| Liver | 2 | 7906 | 3953 | |
| Lung | 2 | 559 | 280 | |
| Skeletal muscle | 2 | 12 932 | 6466 | |
| Skin | 2 | 9069 | 4535 | |
| Stomach | 2 | 7104 | 3552 | |
| Thyroid gland | 2 | 4190 | 2095 | |
| Tongue | 2 | 5024 | 2512 | |
| Umbilical cord | 2 | 1665 | 833 | |
| Uterus | 2 | 13 313 | 6657 | |
| Mouse | Brain | 4 | 5102 | 1276 |
| Heart | 4 | 797 | 199 | |
| Kidney | 1 | 624 | 624 | |
| Liver | 4 | 982 | 246 | |
| Lung | 4 | 1066 | 267 | |
| Skin | 1 | 806 | 806 | |
| Spleen | 1 | 546 | 546 | |
| Testis | 4 | 5374 | 1344 | |
| Thymus | 1 | 683 | 683 |
Tissue-specific circRNAs in the human and mouse genomes
| Species . | Tissue type . | Number of samples . | Number of TS circRNAs . | Average number of TS circRNAs per sample . |
|---|---|---|---|---|
| Human adult | Adipose | 8 | 11 020 | 1378 |
| Adrenal | 2 | 3594 | 1797 | |
| Blood vessel | 2 | 4464 | 2232 | |
| Esophagogastric | 4 | 8876 | 2219 | |
| Esophagus | 8 | 19 503 | 2438 | |
| Female gonad | 2 | 5455 | 2728 | |
| Heart | 5 | 15 278 | 3056 | |
| Intestine | 6 | 15 505 | 2584 | |
| Liver | 4 | 21 536 | 5384 | |
| Lung | 4 | 5230 | 1308 | |
| Mammary gland | 3 | 9193 | 3064 | |
| Pancreas | 2 | 1727 | 864 | |
| Skeletal muscle | 3 | 5546 | 1849 | |
| Skin | 2 | 3968 | 1984 | |
| Thyroid gland | 1 | 3599 | 3599 | |
| Tibial nerve | 4 | 6187 | 1547 | |
| Human fetus | Adrenal | 2 | 358 | 179 |
| Brain | 10 | 89 137 | 8914 | |
| Eye | 2 | 10 495 | 5248 | |
| Heart | 3 | 1608 | 536 | |
| Intestine | 2 | 390 | 195 | |
| Kidney | 2 | 319 | 160 | |
| Liver | 2 | 7906 | 3953 | |
| Lung | 2 | 559 | 280 | |
| Skeletal muscle | 2 | 12 932 | 6466 | |
| Skin | 2 | 9069 | 4535 | |
| Stomach | 2 | 7104 | 3552 | |
| Thyroid gland | 2 | 4190 | 2095 | |
| Tongue | 2 | 5024 | 2512 | |
| Umbilical cord | 2 | 1665 | 833 | |
| Uterus | 2 | 13 313 | 6657 | |
| Mouse | Brain | 4 | 5102 | 1276 |
| Heart | 4 | 797 | 199 | |
| Kidney | 1 | 624 | 624 | |
| Liver | 4 | 982 | 246 | |
| Lung | 4 | 1066 | 267 | |
| Skin | 1 | 806 | 806 | |
| Spleen | 1 | 546 | 546 | |
| Testis | 4 | 5374 | 1344 | |
| Thymus | 1 | 683 | 683 |
| Species . | Tissue type . | Number of samples . | Number of TS circRNAs . | Average number of TS circRNAs per sample . |
|---|---|---|---|---|
| Human adult | Adipose | 8 | 11 020 | 1378 |
| Adrenal | 2 | 3594 | 1797 | |
| Blood vessel | 2 | 4464 | 2232 | |
| Esophagogastric | 4 | 8876 | 2219 | |
| Esophagus | 8 | 19 503 | 2438 | |
| Female gonad | 2 | 5455 | 2728 | |
| Heart | 5 | 15 278 | 3056 | |
| Intestine | 6 | 15 505 | 2584 | |
| Liver | 4 | 21 536 | 5384 | |
| Lung | 4 | 5230 | 1308 | |
| Mammary gland | 3 | 9193 | 3064 | |
| Pancreas | 2 | 1727 | 864 | |
| Skeletal muscle | 3 | 5546 | 1849 | |
| Skin | 2 | 3968 | 1984 | |
| Thyroid gland | 1 | 3599 | 3599 | |
| Tibial nerve | 4 | 6187 | 1547 | |
| Human fetus | Adrenal | 2 | 358 | 179 |
| Brain | 10 | 89 137 | 8914 | |
| Eye | 2 | 10 495 | 5248 | |
| Heart | 3 | 1608 | 536 | |
| Intestine | 2 | 390 | 195 | |
| Kidney | 2 | 319 | 160 | |
| Liver | 2 | 7906 | 3953 | |
| Lung | 2 | 559 | 280 | |
| Skeletal muscle | 2 | 12 932 | 6466 | |
| Skin | 2 | 9069 | 4535 | |
| Stomach | 2 | 7104 | 3552 | |
| Thyroid gland | 2 | 4190 | 2095 | |
| Tongue | 2 | 5024 | 2512 | |
| Umbilical cord | 2 | 1665 | 833 | |
| Uterus | 2 | 13 313 | 6657 | |
| Mouse | Brain | 4 | 5102 | 1276 |
| Heart | 4 | 797 | 199 | |
| Kidney | 1 | 624 | 624 | |
| Liver | 4 | 982 | 246 | |
| Lung | 4 | 1066 | 267 | |
| Skin | 1 | 806 | 806 | |
| Spleen | 1 | 546 | 546 | |
| Testis | 4 | 5374 | 1344 | |
| Thymus | 1 | 683 | 683 |
Identification of TS circRNAs
According to previous research, we applied three algorithms, CIRI [7, 33], circRNA_finder [34] and find_circ [6], which is the combination with the most efficiency and accuracy [35] to detect the back-splice junction sites of circRNAs. To ensure the high accuracy for the identification of circRNAs, we kept the circRNAs identified by either one of three algorithms and required ≥2 junction reads (Supplementary Figure S1A). To identify TS circRNAs, we selected those circRNAs identified in only one tissue for our further analysis (Supplementary Figure S1B).
Validation of TS circRNAs by reverse transcription polymerase chain reaction
To validate the existence of TS circRNAs, we performed reverse transcription polymerase chain reaction (RT-PCR) in five mouse tissues, including brain, heart, liver, lung and thymus. C57BL/6 mice were obtained from the Wuhan University Center for Animal Experiment/ABSL-3 Laboratory. The protocol was approved by the Committee on the Ethics of Animal Experiments of Wuhan University (China) (Permit Number: SCXK 2008-0004). Total RNA from five mouse tissues was prepared by the TRIzol reagent (Invitrogen, Cat 15596026) according to the manufacturer’s instructions. All the RNAs were digested by RNase-free DNase I and purified, and 3 μg of RNA was used as template for RT using RevertAid First Strand cDNA Synthesis Kit (Thermo Fermentas, Cat K1622). PCR was performed in a 20 ul reaction mix using 2XPCR Reagent (Tiangen, KT207-1). All primers were listed in Supplementary Table S1.
Characterization of genomic location and conservation of TS circRNA
We constructed gene annotation based on human hg19 and mouse mm9 genome version by integrating REFSEQ [36] (http://www.ncbi.nlm.nih.gov/refseq/), UCSC [37] (http://genome.ucsc.edu) and GENCODE LncRNA [38] (http://www.gencodegenes.org/) database. We obtained 96 169 and 40 733 transcripts in human and mouse, respectively. We then mapped the circRNA donator/acceptor sites to the annotated gene regions classified snoRNAs into exonic, intronic and intergenic groups. By integrating the long noncoding RNA (lncRNA) from REFSEQ, UCSC and GENCODE annotation, we also classified circRNAs into mRNA and lncRNA groups. The coordinates of backsplice sites of TS circRNA were converted between human and mouse using liftOver from UCSC (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Then the sites were compared to determine the conservation of circRNAs. We allowed 2 bp window while comparing the backsplice sites as previously described [2]. To normalize the abundance of TS circRNAs, we defined a customized SRPTM [number of circular reads/number of mapped reads (units in trillion)/read length], which was adopted from SRPBM [number of circular reads/number of mapped reads (units in billion)/read length] from previous work [39].
Functional enrichment analysis by gene ontology
The TS circRNA derived genes were used for the functional enrichment analysis. Those highly expressed TS circRNAs (SRPTM > 8.5) were selected. The enrichment analysis was performed in Gene Ontology (GO) biological processes by DAVID Bioinformatics Resources v6.7 [40]. Fisher’s Exact test was performed to determine the P-value. The most enriched pathways in human and mouse tissues were plotted.
Identification of miRNA response elements and protein binding sites on TS circRNAs
The regions surrounding the backsplice site were often used to predict the potential miRNA or protein binding. According to previous research, we selected a 100 bp window (±50 bp) as previously described [41] surrounding the backsplice site to scan the potential miRNA response elements (MRE) using TargetScan [42]. We scanned the junction region for the miRNA seeds: 7mer-m8, 7mer-1a and 8mer as previously described [43].
CLIP-Seq data were downloaded from STARBASE [44] including protein binding sites of 37 RBPs. Similar to MRE analysis, we also scanned the junction regions (±50 bp surrounding backsplice sites) for RBP sites.
Construction of TS circRNA database
All of the data were organized into a set of relational MySQL tables. Customized Java and PHP scripts were used to construct the interface of database. The visualization page displayed the coordinates of each circRNA. The circRNA reference was constructed by merging the exons of different transcripts from one gene. The coordinates for each circRNA based on human GRCH38 and mouse mm10 genome version were also provided.
Results
Identification and characterization of TS circRNA
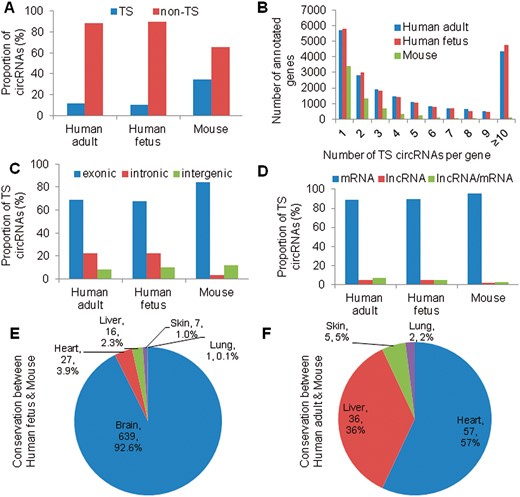
We collected 123 rRNA-depleted RNA-seq data sets from ENCODE and GEO database, which included 16 adult human adult tissues, 15 fetal human fetal tissues and 9 mouse tissues (Table 1). We identified 140 681, 164 069 and 15 980 TS circRNAs in 16 human adult tissues, 15 human fetal tissues, 9 mouse tissues, respectively (Table 1). For all circRNAs, we observed 1 143 367 circRNAs are overlapped between human adult (96.5% of 1 184 752) and fetal tissues (72.3% of 1 580 940). Among these, 11.9% (140 681 of 1 184 752) of circRNAs in human adult, 10.4% (164 069 of 1 580 940) of circRNAs in human fetus and 34.3% (15 980 of 46 579) in mouse are tissue specific (Figure 1A). We then examined the distribution of TS circRNA in annotated genes and found that majority of the genes contained <3 TS circRNAs (84.8% in mouse, 51.9% in human adult and 52.3% in human fetus), suggesting that only a small fraction of genes produced abundant TS circRNAs (≥3) (Figure 1B).
Comprehensive characterization of TS circRNAs in human, fetal and adult, and mouse tissues. (A) Percentage of TS and non-TS circRNAs. (B) Number of TS circRNAs per gene. (C) Distribution of TS circRNA in exonic, intronic and intergenic regions. (D) Distribution of TS circRNA generated from mRNA and/or lncRNA. (E) Number of conserved circRNAs in same tissues between fetal human and mouse. (F) Number of conserved circRNAs in same tissues between adult human and mouse. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
Previous work revealed the variant origins of circRNAs in genomic regions [6, 39]. To further examine the origins of TS circRNAs, we examined the genomic location of TS circRNAs in our database. We found that TS circRNAs origination was different in human and mouse. For example, in mouse, 84.3%, 3.5% and 12.2% of TS circRNAs were located in exonic, intronic and intergenic regions, respectively. While in the human adult, 69.2%, 22.6% and 8.2% were located in exonic, intronic and intergenic regions, respectively. In the human fetus, 67.8%, 22.3% and 9.9% were located in exonic, intronic and intergenic regions, respectively (Figure 1C).
Moreover, circRNAs were reported to be associated to the pre-mRNA splicing [8], and therefore, most of circRNAs were generated from protein coding genes [39]. Here, we identified that 88.7%, 4.5% and 6.8% of TS circRNAs were derived from mRNA, lncRNA and overlapped regions of mRNA and lncRNA, respectively, in human adult tissues. Similarly, we identified that 89.8%, 5.2% and 5.0% of TS circRNAs were derived from mRNA, lncRNA and overlapped regions of mRNA and lncRNA, respectively, in human fetal tissues. Meanwhile, we identified that 95.8%, 1.9% and 2.3% of TS circRNAs were derived from mRNA, lncRNA and overlapped regions of mRNA and lncRNA, respectively, in mouse tissues (Figure 1D). This result revealed that mRNA-derived circRNAs are much more than noncoding RNA-derived circRNAs, which is consistent to previous work [6].
We compared the backsplice sites between human and mouse tissues and checked the conservation of circRNAs across the same tissues. We observed that 639, 27, 16, 7 and 1 circRNAs were conserved between mouse and fetal human brain, heart, liver, skin, lung (Figure 1E). A total of 57, 36, 5 and 2 circRNAs were conserved between mouse and adult human heart, liver, skin and lung (Figure 1F). The conserved circRNAs in these tissues may indicate their important functions in organ development.
Abundance of TS circRNAs in different tissues
We then examined the expression abundance of TS circRNAs in these data sets. We observed the obvious specificity of circRNAs among tissues. In adult human, we observed high abundance of TS circRNAs in the esophagus, heart, intestine and liver (Figure 2A). In fetal human, brain, skeletal muscle and uterus have abundant TS circRNAs (Figure 2B). In mouse available tissues, we found brain and testis have the highest abundance of TS circRNAs (Figure 2C). For example, we found protein-coding gene CORIN (corin, serine peptidase), which is a heart-specific Serine Proteinase, generated 40 heart-specific circRNAs. Among these, 25 TS circRNAs were adult-specific, suggesting circRNAs from this gene are specific to the functions of adult heart (Supplementary Figure S2A). Another example is ALB (albumin), which functions primarily as a carrier protein for steroids, fatty acids and thyroid hormones. It also plays a role in stabilizing extracellular fluid volume, generating 160 liver-specific TS circRNAs. Among these, 95 were adult liver specific, and 33 were fetal liver specific (Supplementary Figure S2B). This suggested that TS circRNAs may be associated with the function in organ development and differentiation.
Abundance of TS circRNAs across different tissues. (A) 16 adult human tissues. (B) 15 fetal human tissues. (C) 9 mouse tissues. Abundance of circRNAs is represented by log2 of SRPTM [number of circular reads/number of mapped reads (units in trillion)/read length] with the color depth and column width representing the number of TS circRNAs in different tissues. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
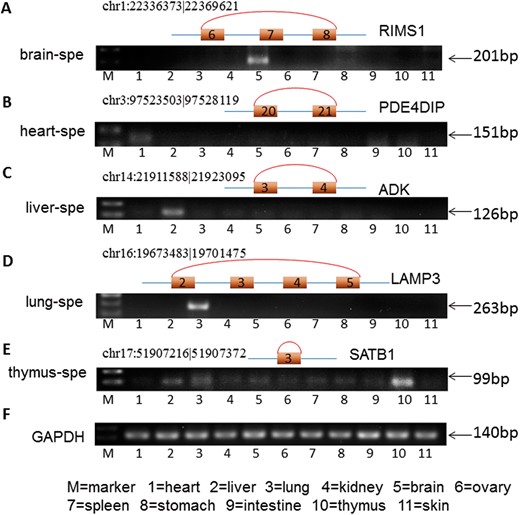
To validate the existence of TS circRNA, we perform RT-PCR experiment based on mouse tissues. TS circRNAs from five mouse tissues including the brain, heart, liver, lung and thymus were validated. These TS circRNAs are generated from exons of five protein-coding genes: RIMS1 in brain, PDE4DIP in heart, ADK in liver, LAMP3 in lung and SATB1 in thymus separately (Figure 3). The RT-PCR results confirmed the high accuracy of our algorithms to identify TS circRNAs.
Validation of TS circRNAs by RT-PCR in mouse tissues. (A) RIMS1 in brain. (B) PDE4IP in heart. (C) ADK in liver. (D) LAMP3 in lung. (E) SATB1 in thymus. GAPDH was used to control the quality of total RNA. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
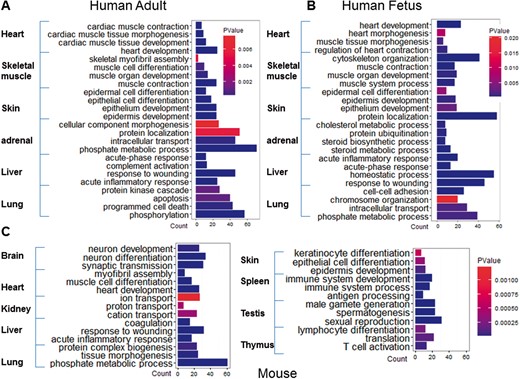
To further explore the potential function of TS circRNAs in organ development or differentiation, we selected TS circRNAs-derived genes to perform the GO enrichment. The results indicated that TS circRNAs were enriched in the pathway of special tissue development. For example, the heart-specific circRNAs were enriched in the process of cardiac muscle tissue development in both adult and fetus (Figure 4A and B). In consistent with this, TS circRNAs in mouse were also enriched in tissue development and differentiation (Figure 4C).
Pathway clustering of TS circRNAs in overlapped adult and fetal human tissues. GO biological process were identified for TS circRNAs-derived linear mRNA in five human tissues (heart, skeletal muscle, skin, adrenal, liver and lung), and 9 mouse tissues (brain, heart, kidney, liver, lung, skin, spleen, testis, and thymus). (A) Pathway clusters in adult human tissues. (B) Pathway clusters in fetal human tissues. (C) Pathway clustering in mouse tissues. Color depth represents the P-value of each process calculated by Fisher’s Exact test, and column length represents the count of TS circRNAs. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
Potential functions through microRNA and protein binding
CircRNAs were mainly reported to exert the function through binding microRNAs, which are called miRNA sponges [12–14, 45]. To reveal the potential functions of TS circRNA, we investigated the MRE and identified 264 728 MREs in 15 980 mouse TS circRNAs (on average 17 MREs per TS circRNA), 3 047 871 MREs in 140 681 human adult TS circRNAs (on average 22 MREs per TS circRNA) and 3 482 558 MREs in 164 069 human fetal TS circRNAs (on average 21 MREs per TS circRNA). For example, miR-7 response elements were frequently detected in adult adipose, heart and skeletal muscle TS circRNAs, which suggested miR-7 functions as a modulator of several oncogenes and associated with various cancers [12]. All MRE prediction results were deposited in TSCD database.
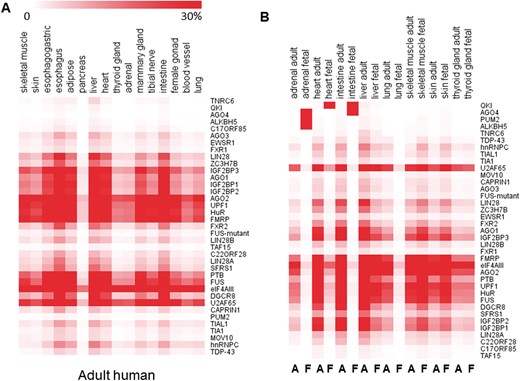
Moreover, circRNAs can also bind to proteins directly to exert more functions [16, 17], and protein binding also can affect the biogenesis of circRNAs [11]. To reveal the potential function with circRNA binding proteins, we downloaded CLIP-Seq data sets from STARBASE including 37 RBPs and examined the RBP binding sites in the junction regions (±50 bp surrounding junction sites) for TS circRNAs. The distribution of 37 RBPs were different between tissues (Figure 5A, Supplementary Figure S3A and B). For example, we observed that the Argonaute family member AGO1 and AGO2 displayed abundant binding sites in TS circRNAs in most of 16 adult tissues while AGO3 and AGO4 displayed few binding sites (Figure 5A). By comparing the adult and fetal human tissues, we observed that TS circRNAs could bind to different RBPs at the different developmental stage. For example, AGO4 arise more in fetal than in adult intestine. Reversely, AGO1 and AGO2 arise more in adult than in fetus intestine (Figure 5B). Taken together, our results revealed different binding potency of RBPs in those TS circRNAs among development stages.
Potential RBPs in junction regions of TS circRNAs. (A) Potential RBP sites in TS circRNAs of human adult tissues. (B) Potential RBP sites in TS circRNAs in both human adult and fetal tissues (A = Adult, F = Fetus). Color depth represents the percentage of number of RBP sites. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
A TS circRNA database TSCD
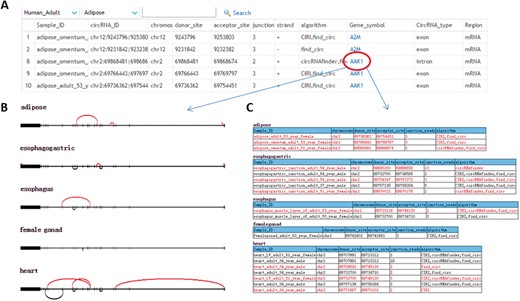
We have identified 302 853 TS circRNA, as well as the potential MREs and RBP sites, and we also revealed the potential functions of TS circRNAs. To bridge the gap between large information and biologist, we construct a comprehensive database, named TSCD. Users can easily browse TSCD content through browser page. The users can view the TS circRNA by selecting tissues from human adult, human fetus and mouse—specifically, 26 individual tissues, including adipose, adrenal, blood vessel, brain, esophagogastric, esophagus, eye, female gonad, heart, intestine, kidney, liver, lung, mammary gland, pancreas, skeletal muscle, skin, spleen, stomach, testis, thymus, thyroid gland, tibial nerve, tongue, umbilical cord and uterus. The users can also view the comprehensive information, such as tissue category, circRNA ID, coordinates of backsplice sites, genomic locations, junction reads, strand information, genomic spanning length, gene annotation and MRE/RBP sites (Figure 6A). More importantly, users can visualize the details of TS circRNA through the gene symbol link. The nonspecific circRNAs were displayed as black arc, while the TS circRNA were displayed as red arc. The annotated exon and intron of reference transcripts were displayed in the following panel. If the reference genes have multiple transcripts, all the transcripts were displayed (Figure 6B). If the circRNA is generated from multiple genes, the exon structures of all related genes were displayed to better illustrate the biogenesis of circRNAs. TSCD provided the tables including all precise coordinates of each backsplice of circRNA across different tissues (Figure 6C).
Web interface of TSCD. (A) The index page allows the user to easily query the information of TS circRNAs by chromosome, start and end site, junction read, conservation, genomic location, etc. (B) Illustration of circRNAs generated from annotated genes with the exon structures. Backsplices of circRNA were represented by arc. TS circRNAs were displayed in red arcs and non-TS circRNAs were displayed in black arcs. (C) Detailed information for tissue and coordinates of circRNAs from panel B. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
Furthermore, TSCD also provided the additional pages to benefit the research community: (1) the Browser-hg38|mm10 page, which displayed coordinates for each circRNA based on the latest genome version, including GRCH38 and mm10; (2) comparison page, which allowed the users to compare circRNAs among different tissues; (3) download page, which allowed the users to batch download TS circRNAs from all tissues and the customized Perl script to identify the TS circRNAs from their own RNA-seq data.
Conclusions
As circRNAs are attracting more attention in transcriptome research, we are exploring the global features of TS circRNAs in development and organ differentiation. Based on the major types of circRNA (Supplementary Figure S4), we identified >300 000 of TS circRNAs in different tissues. Our analysis indicated that TS circRNAs were mainly derived from exons, but may also derived from intron or intergenic regions. The majority of TS circRNAs were generated from protein coding genes, which suggested these circRNAs may be associated to mRNA translation or be a backup of mRNA. Among all circRNAs, 10.4% of human circRNAs and 34.3% of mouse circRNAs are tissue specific, which suggested the association of TS circRNAs to tissue development.
We also observed uneven distribution of TS circRNAs across different tissues. There are more TS circRNAs expressed in brain (89 137 were identified in fetal brain), and this might be owing to the complexity of neuronal activity in brain [23]. We then performed the functional enrichment analysis for TS circRNAs, and our results suggested that TS circRNAs are largely associated with tissue development and differentiation. To understand the potential functions of TS circRNAs, we identified a significant number of miRNA binding elements (MRE) and RBP binding sites.
Finally, to benefit the community, we constructed a database, TSCD. This includes information for TS circRNAs, such as origination of genomic location, conservation across species and visualization of exons structures. TSCD provides a new integrating view to explore the TS circRNAs in human and mouse tissues. We will continually update TSCD to include the latest available RNA-seq data sets, especially those RNA-seq more appropriate to identify circRNAs, such as RNase R treatment.
The first global analysis of tissue-specific circRNAs.
Characterize genomic features of tissue-specific circRNAs.
Predict functions of tissue-specific circRNAs.
Construct a comprehensive database for tissue-specific circRNAs.
Siyu Xia is a master student at School of Basic Medical Sciences, Wuhan University.
Jing Feng is an associate professor at International School of Software, Wuhan University.
Lijun Lei is a master student at School of Basic Medical Sciences, Wuhan University.
Jun Hu is a postdoc fellow at School of Basic Medical Sciences, Wuhan University.
Linjian Xia is a master student at School of Basic Medical Sciences, Wuhan University.
Jun Wang is a master student at School of Basic Medical Sciences, Wuhan University.
Yu Xiang is a postdoc fellow at Department of Biochemistry and Molecular Biology, The University of Texas Health Science Center at Houston McGovern Medical School.
Lingjun Liu is a master student at School of Basic Medical Sciences, Wuhan University.
Shan Zhong is a professor at School of Basic Medical Sciences, Wuhan University.
Leng Han is an assistant professor and CPRIT scholar at Department of Biochemistry and Molecular Biology, The University of Texas Health Science Center at Houston McGovern Medical School. He is an expert for high-throughput data mining, especially for RNA sequencing.
Chunjiang He is an associate professor at School of Basic Medical Sciences, Wuhan University. He is an expert for identification and functional characterization of noncoding RNAs.
Acknowledgments
The authors thank Wuhan University and The University of Texas Health Science Center at Houston for financial support to this research. We also thank Wuhan University Center for Animal Experiment/ABSL-3 Laboratory for providing the experimental mice. We thank Martha Czernuszenko for proofreading.
Funding
National Natural Science Foundation of China (81500140 to C.H.); Natural Science Foundation of Hubei Province, China (2015CFB170 to C.H.); the Fundamental Research Funds for the Central Universities of China (2042015kf0018 to C.H.); Cancer Prevention & Research Institute of Texas (RR150085 to L.H.).
References
Author notes
Siyu Xia and Jing Feng authors contributed equally.


![Abundance of TS circRNAs across different tissues. (A) 16 adult human tissues. (B) 15 fetal human tissues. (C) 9 mouse tissues. Abundance of circRNAs is represented by log2 of SRPTM [number of circular reads/number of mapped reads (units in trillion)/read length] with the color depth and column width representing the number of TS circRNAs in different tissues. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/bib/18/6/10.1093_bib_bbw081/2/m_bbw081f2.jpeg?Expires=1716364465&Signature=JN0hKm6QiCFcS7yMb7Ru7x17XaN~71oloHk5Ai77w8BvzWN3ZGe2vAueoKZu~xYaSlTF6EsRCpnaEuSsWDhESG-uin-wNsOSlgwjpDhrIXB1D0DT9OLSwjLKb8h16AavTN5E442orhbMbVGhbJ8FghHf5QhDtSfrjkZ-YsdnVMpKCpmGUsYrtwM~-rD4v1XzC2rAxIcGPnPAmlXvrO7IlWh7pjdmEbOj7cf8orOcmaBR9EUpi1oiH6h1icveM9R7DH08YFy3HuU~6rDbkIT39k6ppNFxHS9PE33Bk0HJKml0f4XFw-2yyJT6GyKKVs7PvExO2~wCK6JZP5AHqP9ecA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)