Abstract
Spatio-temporal specific long noncoding RNAs (lncRNAs) play important regulatory roles not only in the growth and development of the brain but also in the occurrence and development of neurologic diseases. Generally, the occurrence of neurologic diseases is accompanied by neuroinflammation. Elucidation of the regulatory mechanisms of lncRNAs on neuroinflammation is helpful for the clinical treatment of neurologic diseases. This paper focuses on recent findings on the regulatory effect of lncRNAs on neuroinflammatory diseases and selects 10 lncRNAs that have been intensively studied to analyze their mechanism action. The clinical treatment status of lncRNAs as drug targets is also reviewed.
SIGNIFICANCE STATEMENT Gene therapies such as clustered regularly interspaced short palindrome repeats technology, antisense RNA technology, and RNAi technology are gradually applied in clinical treatment, and the development of technology is based on a large number of basic research investigations. This paper focuses on the mechanisms of lncRNAs regulation of neuroinflammation, elucidates the beneficial or harmful effects of lncRNAs in neurosystemic diseases, and provides theoretical bases for lncRNAs as drug targets.
Introduction
Up to 98% of human transcribed genes do not encode proteins but are transcribed as ncRNAs. A noncoding RNA with a length greater than 200 nucleotides is called a long noncoding RNA (lncRNA) (Hüttenhofer et al., 2005). LncRNA is distributed in many tissues and organs of the human body, but the expression level of lncRNA in vivo is lower than that of the coding genes. It is worth noting that lncRNAs are spatially and temporally specific (Herriges et al., 2014). The time specificity of lncRNA indicates that the expression of lncRNA is different in different stages of growth and development of the body or different stages of the occurrence and development of diseases. The spatial specificity of lncRNA indicates that the level of lncRNA is different in different tissues, and highly expressed lncRNAs in tissues often play important roles. Spatiotemporal-specific lncRNAs are biomarkers integrating sensitivity and specificity, which can be used as screening markers, diagnosis markers, and prognosis markers in diseases (Lei et al., 2018). On the one hand, transcription factors, RNA-binding proteins, and micro RNAs (miRNAs) target lncRNAs to regulate the expression of target lncRNAs (Yang et al., 2019a; Li et al., 2020b). On the other hand, by binding RNA, DNA, and protein, lncRNA regulates related genes at multiple molecular levels, such as epigenetic regulation, transcriptional regulation, and post-transcriptional regulation. For example, lncRNA recruits DNA methyltransferase to regulate the methylation modification of target genes; lncRNA binds to transcription factors to form a complex to regulate the transcription of target genes; and lncRNA binds to 3′-UTR of miRNA to regulate downstream mRNA (Zhou et al., 2015a; Jia et al., 2019; Ma et al., 2019).
Neurologic diseases are physiologic imbalances, which can be perceived as abnormal levels of body components under the joint action of the living environment and genetic code. Neurosystemic diseases, including ischemic stroke, neuropathic pain, and neurodegenerative disorders, have complex pathologic processes and persistent neurologic damage, making their diagnosis, treatment, and prognosis difficult. In-depth studies have revealed that not only is neuroinflammation a common pathologic process of neurosystemic diseases but also that chronic persistent neuroinflammation may cause secondary damage (Fabisiak and Patel, 2022; Jiang et al., 2022; Zhang et al., 2022a). The main processes of neuroinflammation include the actions of microglia and astrocyte cells in the central immune system and the behaviors of macrophages and white blood cells in the peripheral immune system (Carson et al., 2006; Singh, 2022). The development of neuroinflammation is related to immune system cell phenotypes change, inflammatory factors release, inflammasome activation, and signal pathways activation, which is accompanied by the destruction of the blood-brain barrier and the penetration of cerebrospinal fluid (Takata et al., 2021).
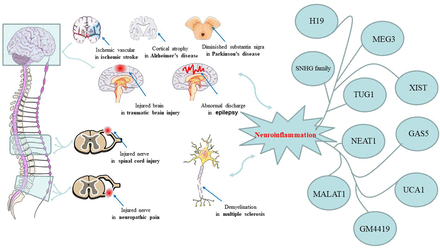
As biomarkers and regulators, lncRNAs take part in neurosystemic diseases, due to the easy availability of blood and the high sensitivity and specificity of lncRNAs. Blood lncRNAs are ideal biomarkers for the diagnosis and prognosis of diseases (Badowski et al., 2022). As regulatory factors, lncRNAs regulate cell proliferation, differentiation, invasion, migration, inflammation, and vascular formation and are considered as potential drug targets (Li et al., 2016). It should be noted that lncRNA plays a pro-inflammatory or an anti-inflammatory role in inflammation, regulating the activation of glial cells, the release of inflammatory factors such as IL-6, cyclooxygenase-2 (COX-2), tumor necrosis factor alpha (TNF-α), and the activation of absent in melanoma 2 (AIM2), Nlr family pyrin domain containing 3 (NLRP3) inflammasome, nuclear factor kappa light chain enhancer of activated B cells (NF-κB), phosphatidylinositol-3-kinase/Ak strain transforming; (PI3K/AKT), Janus kinase/signal transducer and activator of transcription (JAK/STAT) signal pathways (Cao et al., 2018; Han et al., 2018; Zhou et al., 2018a; Liang et al., 2020). Different lncRNAs may play similar roles in neuroinflammation associated with the same disease, and the same lncRNA may play different roles in neuroinflammation associated with different diseases. At present, some articles have reviewed the role of lncRNA in neurosystemic diseases (Chen et al., 2021; Ebrahimi and Golestani, 2022; Jiang et al., 2022), but there is no article on the comparison of the role of the same lncRNA in neuroinflammation in different diseases. In this paper, 10 lncRNAs related to neuroinflammation and their roles in different neurosystemic diseases are highlighted, and the current application of lncRNAs that act as drug targets in clinical treatment is reviewed (Fig. 1, Table 1).
Common neurosystemic diseases and lncRNAs associated with neuroinflammation. Some common neurosystemic diseases include ischemic stroke, Alzheimer's disease, Parkinson's disease, traumatic brain injury, epilepsy, multiple sclerosis, spinal cord ischemia, neuropathic pain, and multiple sclerosis. Each disease has its own unique pathologic features, but neuroinflammation is the common pathologic process. Reducing the production of neuroinflammation in a way that improves the brain environment, protects nerve cells, and alleviates the progression of the disease. Accumulated studies have confirmed that lncRNAs play pro-inflammatory or anti-inflammatory roles in the regulation of neuroinflammation. Furthermore, lncRNAs including MALAT1, NEAT1, TUG1, SNHG family, H19, MEG3, XIST, GAS5, UCA1, and GM4419 have been found to alleviate more than one neurosystemic disease by regulating neuroinflammation.
Functional characterizations of lncRNAs in neuroinflammation-related diseases.
Expression of 10 lncRNAs (MALAT1, NEAT1, TUG1, SNHG family, H19, MEG3, XIST, GAS5, UCA1, and GM4419) in different neurosystemic disease (ischemic stroke, Alzheimer's disease, Parkinson's disease, traumatic brain injury, epilepsy, multiple sclerosis, spinal cord ischemia, neuropathic pain, and multiple sclerosis) models and biologic functions of lncRNAs by regulating downstream targets.
MALAT1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is located at chromosome 11q13.1. It is universally expressed in human tissues including bone marrow, thyroid, and prostate. Transcription factor STAT3, Yes-associated protein 1, and Kruppel like factor 4 promote the expression of MALAT1 through targeting promoter. RNA such as miR-146b-5p, miR-9, and lncRNA differentiation antagonizing non-protein coding RNA could regulate the level of MALAT1. Furthermore, disruptor of telomeric silencing 1-like protein also directly binds to MALAT1 (Leucci et al., 2013; Duan et al., 2019; Sun et al., 2019; Peng et al., 2021; Xiong et al., 2021; Jing et al., 2022; Yang et al., 2018b). More significantly, the two main post-transcriptional modifications of MALAT1 are N6-methyladenosine and 5-methylcytosine modification (Squires et al., 2012; Liu et al., 2013). MALAT1 interacts with RNA, DNA, and protein to realize its biologic function. For example, MALAT1 is combined with miRNA, which jointly regulates downstream mRNA. Binding to serine/arginine-rich family splicing factors or RNA binding proteins to show the ability of alternative splicing and transcriptional regulation (El Bassit et al., 2017; Xie et al., 2017; Scherer et al., 2020). MALAT1 plays an important regulatory role in physiologic and pathologic processes. In a physiologic state, MALAT1 plays a regulatory role in cell proliferation, differentiation, migration, epithelial-mesenchymal transition, autophagy, apoptosis, and so on (Cheng et al., 2019; Luo et al., 2019; Bao et al., 2020; He et al., 2020; Pi et al., 2022; Zang et al., 2022).
MALAT1 functions as a marker and regulator in disease states. As a screening marker, single nucleotide polymorphisms analysis of MALAT1 showed that MALAT1 was associated with cancer and immune disease susceptibility (Chen et al., 2020; Mao et al., 2021). MALAT1 as a diagnostic marker can be used to diagnose atherosclerotic cardiovascular disease, lung cancer, and breast cancer (Zhao et al., 2020b; Liu et al., 2021b). When combined with other biomarkers for the diagnosis of disease, the sensitivity, and specificity of diagnosis are improved; for example, MALAT1 interacts with miR-125b as a coronary heart disease biomarker, MALAT1-H19/miR-19b-3p axis as a diabetic neuropathy biomarker (Lv et al., 2021a; Rajabinejad et al., 2022), and MALAT1 as a prognostic marker. Overexpression of MALAT1 was associated with a dismal prognosis, manifested in glioblastoma, lung cancer, and other numerous malignancies (Li et al., 2018a).
MALAT1 acts as a detrimental factor or protective factor in the occurrence and development of different diseases. Therefore, strengthening the protective effect and decreasing the dangerous effect of MALAT1 by regulating the level of MALAT1 may reverse the disease process, making it a potential target for disease treatment (Abdulle et al., 2019; Yang et al., 2020). In the study of MALAT1 in acute spinal cord injury, MALAT1 has been found to be associated with neuroinflammation. Subsequent researchers have shown that MALAT1 also plays a role in the neuroinflammation that accompanies neuropathic pain, multiple sclerosis, Alzheimer’s disease, traumatic brain injury, Parkinson’s disease, and cerebral ischemic stroke. MALAT1 was up-regulated in a chronic constriction injury rat model of neuropathic pain, and the pain threshold assessment and expression of IL-6, IL-1β, TNF-α, and COX-2 showed that MALAT1 promoted the development of neuropathic pain and neuroinflammation. The current study found three different mechanisms to achieve this effect(Chen et al., 2019b; Ma et al., 2020b; Wu et al., 2020b). Ma et al. confirmed that MALAT1 and miR-129-5p are highly expressed in neuropathic pain, and together with the opposite expression of HMGB1 [HMGB1 protein is a kind of protein that usually binds with cytokines TNF-α, IL-6, and IL-1β to trigger inflammatory response and plays an important role in the induction of inflammation and autophagy (Mori et al., 2018)], the competing endogenous RNA (ceRNA) network formed by the three factors regulates the disease process (Ma et al., 2020b). Chen et al. showed that MALAT1 played a pro-inflammatory role by targeting the miR-206/ZEB2 signal axis (Chen et al., 2019b). Wu et al. verified the role of MALAT1/miR‐154-4p/AQP9 axis in neuropathic inflammation (Wu et al., 2020b). The pro-inflammatory effect of MALAT1 is also reflected in Parkinson’s disease and acute spinal cord injury. Cai et al. showed that MALAT1 down-regulation improved the exercise ability of MPTP-treated C57BL/6 mice. The C57BL/6 mice model treated with MPTP is a valuable model of Parkinson’s disease. In LPS/ATP-pretreated BV2 microglia cells, the ability of MALAT1 to trigger neuronal injury is due to the recruitment of EZH2 to act on the promoter of NRF2 to achieve negative regulation of NRF2, thus activating NLRP3-mediated inflammasome and increasing reactive oxygen species level (Cai et al., 2020b). Zhou et al. showed that MALAT1 down-regulation can inhibit the progression of acute spinal cord injury, while miR-199b inhibitor can induce its production, suggesting that MALAT1 and miR-199b are negatively correlated, which regulates the IKKβ/NF-κB signaling pathway (Zhou et al., 2018a). The IKKβ/NF-κB signal pathway is a classic pathway regulating inflammation that plays a pro-inflammatory role in multiple sclerosis, ischemic stroke, and epilepsy. This pathway also regulates the apoptosis of neurons and the formation of glial scars in spinal cord injury ( Zhou et al., 2016; Babkina et al., 2021).
The anti-inflammatory effect of MALAT1 is reflected in multiple sclerosis, Alzheimer’s disease, ischemic stroke, and traumatic brain injury. In a study related to Alzheimer’s disease, MALAT1 regulates the expression levels of PTGS2, cyclin-dependent kinase 5 (CDK5), and FOXQ1 by regulating miR-125b, which stimulates neurite outgrowth and inhibits neuron apoptosis and neuroinflammation (Ma et al., 2019). The regulation of miR-125b by MALAT1 shows that lncRNA can down-regulate neurotoxic miRNA, making it play a neuroprotective role. MiR-125b aggravates the process of Alzheimer’s disease, which is reflected in down-regulating sphingosine kinase 1 protein expression, making tau hyperphosphorylation, and phosphorylation, neurons apoptosis, and inflammation (Jin et al., 2018c). The effect of MALAT1 antagonizing miR-125b on promoting cell proliferation is also reflected in oral squamous cell carcinoma and bladder cancer (Xie et al., 2017; Chang and Hu, 2018). Ruan et al. found that small molecule Polydatin regulates transcription factor CCAAT/enhancer-binding protein Β binding to the promoter region of MALAT1 and promotes the expression of MALAT1. Up-regulated MALAT1 inhibits inflammatory response and apoptosis by regulating camp response-element binding protein/PGC-1α/PPARγ pathway, ameliorating stroke (Ruan et al., 2019). A study has shown that hASC-derived exosomes contain lncRNAs including MALAT1 as well as proteins. Hasc-derived exosomes tend to migrate to liver, and it is speculated that exosomes may act to reduce the production of spleen immune cells or inhibit the inflammatory response of immune cells, thereby reducing secondary brain damage caused by peripheral immune cells through broken blood-brain barrier (BBB) (Gupta and Pulliam, 2014). In addition, although hASC-derived exosomes rarely migrate into the brain, they have direct neuroprotective effects on nerve cells (El Bassit et al., 2017). HASC-derived exosomes containing MALAT1 inhibit ephrin family gene expression after traumatic brain injury, which can reduce the penetration of peripheral immune cells into the brain by damaging BBB (Patel et al., 2018). Changes in the phenotype of immune cells indicate changes in the function of immune cells. Masoumi et al. found in the multiple sclerosis cell model that the silencing of MALAT1 promotes the differentiation of macrophage and T cell to pro-inflammatory M1 phenotype and TH1/Th17 phenotype, respectively (Masoumi et al., 2019). These results suggest that MALAT1 can be a potential target in multiple sclerosis disease to inhibit the pro-inflammatory effects of macrophages and T cells.
NEAT1
Nuclear paraspeckle assembly transcript 1 (NEAT1) is located at chromosome 11q13.1. NEAT1 is commonly expressed in ovary, prostate, and colon. Transcription factors Yin Yang 1 and STAT3 target the NEAT1 promoter to regulate NEAT1/miR-205-3p/MMP16 and NEAT1/miR-4688/TULP3, respectively (Cai et al., 2020a; Li et al., 2020b). MiR-340-5p targets NEAT1 by regulating NEAT1/HSF1/MMP11 (Gao et al., 2022). THOC4 protein regulates NEAT1 by directly targeting the promoter region or by binding to cleavage and polyadenylation specific factor 6, which can activate NEAT1 (Klec et al., 2022). RNA-binding protein SRSF1 regulates the cell cycle of glioma cells by regulating NEAT1 (Zhou et al., 2019). Under physiologic conditions, NEAT1 is involved in regulating the differentiation of human bone marrow–derived mesenchymal stem cells and human embryonic stem cells. It also participates in the activation of T helper 2 cells (Chen and Carmichael, 2009; Zhang et al., 2019; Huang et al., 2021b).
As an oncogenic gene, NEAT1 is a prognostic marker of cancer such as breast cancer, nasopharyngeal carcinoma, diffuse large B cell lymphoma, and acute lymphoblastic leukemia (Deng et al., 2018; Liu et al., 2019; Pouyanrad et al., 2019; Quan et al., 2020).
As a regulator, it showed pro-inflammatory effects in Parkinson's disease, neuropathic pain, and epilepsy and anti-inflammatory effects in traumatic brain injury. Generally, Parkinson's disease models can be divided into cellular models and animal models. In cellular models, MPP+ is used to recapitulate the disease. In addition, it is more common to use 6-hydroxydopamine and MPTP in animal models. In vivo and in vitro models of Parkinson’s disease, the levels of inflammatory cytokines IL-1β, IL-6, and TNF-α increased. However, NEAT1 knockdown reversed the ascending effect, suggesting a pro-inflammatory effect of NEAT1. Mechanismally, NEAT1 negatively regulated miR-212, which protected SK-N-SH cells that were treated with MPP+. The downstream targets AX1N1 and RAB3IP are combined to form two axes, namely NEAT1/miR-212-3p/AX1N1 and NEAT1/miR-212-5p/RAB3IP (Song et al., 2018; Liu et al., 2020c; Liu et al., 2021d). Previous studies have shown that AX1N1 and RAB3IP were related to cell growth and apoptosis (Guo et al., 2018; Zhou et al., 2018b). However, under the regulation of NEAT1, they could also regulate inflammation to affect disease progression. Xia et al. found that NEAT1 decreased paw withdrawal threshold and paw withdrawal latency of rat model and promoted pro-inflammatory cytokines of spinal cord tissues. When NEAT1 was knocked down by shRNA, the level of miR-381 was increased, while the level of HMGB1 was decreased. Moreover, miR-381 inhibitors can not only reverse the anti-inflammatory effects by the NEAT1 knockdown but can also reverse the improvement of mechanical allodynia and thermal hyperalgesia by HMGB1 knockdown. In conclusion, NEAT1, miR-381, and HMGB1 were involved in neuropathic pain progression and microglial inflammation (Xia et al., 2018). Wan and Yang found that the expression of miR-129-5p and NEAT1 were opposite in CTX-TNA cells treated with IL-1β. CTX-TNA cells treated with IL-1β is a common model of epilepsy that is used extensively for basic research. Wan and Yang emphasized that the expression of Notch1, JAG1, and HES1 was significantly altered under NEAT1/miR-129-5p regulation. Activation of the Notch signal inhibited cell viability and promoted inflammatory cytokines IL-6 and TNF-α (Wan and Yang, 2020). However, there is a controversy about whether NEAT1 acts as a pro-inflammatory gene or an anti-inflammatory factor in ischemic stroke. There are two major cellular models of ischemic stroke: oxygen glucose deprivation of nerve cells (SH-SY5Y cells and N2a cells) and oxygen glucose deprivation of glial cells (BV2 cells and U87 cells). The up-regulation of β-catenin and its downstream c-Myc and CyclinD1 indicates the activation of the Wnt/β-catenin pathway, which is regulated by NEAT1 that up-regulated by Ying Yang 1. Moreover, overexpression and knockdown of NEAT1 resulted in the aggravation and remission of inflammation and apoptosis of microglial cells treated with oxygen-glucose deprivation/reperfusion. However, Ni et al. showed that NEAT1 inhibited AKT/STAT3 signal to promote cell viability and inhibit microglial M1 polarization and cell apoptosis in BV2 cells treated with oxygen-glucose deprivation/reperfusion (Ni et al., 2020). There are two phenotypes of glial cells: the M1 phenotype (pro-inflammatory phenotype) and the M2 phenotype (anti-inflammatory phenotype). The two inconsistent studies on NEAT1 in ischemia/reperfusion may be due to the different cells used and the different oxygen-glucose deprivation/reperfusion time: one was 2H, and the other was 4H. It is well known that glial cells may have different stress responses under different stress conditions. Zhong et al. showed that NEAT1 inhibits the expression of PIDD1, inhibiting the progression of traumatic brain injury. Specifically, knockdown of NEAT1 enhanced the apoptosis of HT22 cells and the release of cytokines IL-1β, TNF-α, and nitrate oxide. However, overexpression of NEAT1 improved the motor function, learning ability, and spatial memory of traumatic brain injury mice. PIDD1 is also known as p53-induced death domain protein 1, which as an effector participates in p53-induced cell death (Berube et al., 2005). But the up-regulation of NEAT1 can be promoted by Bexarotene, which is an RXR-α agonist. RXR-α has been proved to bind to NEAT1 (Zhong et al., 2017).
TUG1
Taurine up-regulated 1 (TUG1) is located at chromosome 22q12.2. It is universally expressed in testis, endometrium, and thyroid. Transcription factor early growth response 1 targets the promoter of TUG1 to up-regulate the level of TUG1, activating EZH2/TIMP2 to promote the progression of adenomyosis (Shi et al., 2019a). Interestingly, a study has shown that miR-1299 and TUG1 can form a feedback loop with the participation of NOTCH3. Specifically, TUG1 is the downstream target of NOTCH3. TUG1 can act as ceRNA and negatively regulate miR-1299. And miR-1299 is a negative regulator of NOTCH3 (Pei et al., 2020b). In osteosarcoma, the FOXM1/TUG1/miR-219a-5p/AKT pathway forms a similar feedback loop (Li et al., 2018f). Another well-known pathway—Notch signal—also can regulate TUG1 to maintain stemness of glioma stem cells (Katsushima et al., 2016). TUG1 regulates angiogenesis and cell differentiation through competitively binding to miRNA under physiologic conditions. For example, TUG1/miR-505-3p/VEGFA axis promotes angiogenesis of HUVECs (Liu et al., 2021c). TUG1/miR-545-3p/cannabinoid receptor 2 axis, TUG1/miR-222-3p/Smad2/7, and TUG1/Lin28A regulates differentiation of osteoblasts (He et al., 2018; Hao et al., 2020; Wu et al., 2020a). In addition, the TUG1/miR-143/FGF1 axis regulates endothelial differentiation of adipose-derived stem cells (Xue et al., 2019).
As a biomarker, TUG1 was associated with a poor prognosis of gastrointestinal, urologic, and hematologic cancers (Huang et al., 2021a). Furthermore, TUG1 was related to gemcitabine resistance in pancreatic ductal adenocarcinoma and chemotherapy resistance in esophageal squamous cell carcinoma (Jiang et al., 2016; Yang et al., 2018a).
As a regulator, TUG1 has been implicated in multiple sclerosis, ischemic stroke, and spinal cord ischemia reperfusion. In these neurosystemic diseases, TUG1 promotes the development of neuroinflammation by activating the NF-κB signal pathway. TUG1 was found to be highly expressed in glial cells and nerve cells of the Parkinson’s disease model. Knockdown of TUG1 in C57BL/6J mice treated with MPTP inhibited the levels of TNF-α, IL-6, and IL-1β. On the other hand, overexpression of TUG1 in MPP+-treated SH-SY5Y cells promoted the development of inflammation. In addition, miR-152-3p was the target of TUG1, which played a protective role in neurons by reducing apoptosis and neuroinflammation. Mechanismally, miR-152-3p could target the tumor suppressor PTEN to regulate SH-5Y5Y cell apoptosis. Therefore, it was concluded that TUG1/miR-152-3p/PTEN played an important role in Parkinson’s disease neuroinflammation (Zhai et al., 2020; Cheng et al., 2021). Yue et al. showed that TUG1 was the ceRNA of miR-9-5p and controlled its downstream target P50. MiR-9-5p bound to the 3′UTR of P50 and negatively regulated the activation of the NF-κB signaling pathway, thereby inhibiting inflammation (Yue et al., 2019). In BV2 cells treated with oxygen-glucose deprivation/reperfusion, TUG1 was up-regulated, while miR-145a-5p was down-regulated. MiR-145a-5p, a downstream target of TUG1, regulated the ratios of p-P65/P65 and p-IkBa/IkBa, inhibiting microglial M1 polarization and neuroinflammation. Therefore, overexpression of TUG1 or knockdown of miR-145a-5p would aggravate stroke-induced neuroinflammation by activating the NFκB pathway (Wang et al., 2019a). In the in vivo experiment of spinal cord ischemia reperfusion (researchers clamp the descending aorta of Sprague–Dawley rats for 14 minutes to simulate spinal cord ischemia, then remove clamps to simulate reperfusion), researchers found inflammation damage (neurologic defects and blood-spinal cord barrier leakage) was mediated by NFκB/IL-1β. Moreover, TUG1 can regulate this signal pathway through miR-29b-1-5P/MTDH and TRIL/TLR4. It is worth mentioning that TLR4-mediated activation of NFκB pathway is the signal of microglia actively participating in inflammatory response (Li et al., 2014). Knockdown of TUG1 by siRNA not only decreased the expression of TLR4 but also inhibited the activation of NFκB pathway mediated by TLR4. TRIL, which was also down-regulated by TUG1-siRNA, also acted on TLR4 to regulate the inflammatory response. These mechanisms explained the phenomenon that down-regulation of TUG1 resulted in decreased release of inflammatory factor IL-1β and decreased number of Iba-1-positive microglia, suggesting a pro-inflammatory role of TUG1 in spinal cord ischemia reperfusion (Jia et al., 2019; Jia et al., 2021).
SNHG family
The SNHG family consists of SNHG1 to SNHG22, a total of 22 members. The roles of family members in cancer have been reviewed, and it can be seen that each member shows different expression patterns and different regulatory mechanisms in cancer (Qin et al., 2020). Studies in recent years have found that family members play regulatory roles in neurologic diseases. In the following, we focus on the roles of SNHG1 and SNHG2 in neurosystemic diseases associated with neuroinflammation.
Small nucleolar RNA host gene 1 (SNHG1) is located at chromosome 11q12.3. It is greatly expressed in bone marrow, ovary, and lymph node. Transcription factor SP1 binds to the promoter region of SNHG1 to regulate transcriptional activity and epilepsy development (Zhao et al., 2020a). In a physiologic state, SNHG1 regulates osteogenic differentiation of human bone marrow stromal cell, fibroblastic cells from the posterior longitudinal ligament, and periodontal ligament stem cells (Li et al., 2020d; Wang et al., 2020a; Zhang et al., 2021a).
SNHG1 is associated with the poor prognosis of osteosarcoma, serous epithelial ovarian cancer, liver cancer, and other cancers (Wang et al., 2018a; Pei et al., 2020a; Zhang et al., 2020a).
As a regulatory factor, SNHG1 plays pro-inflammatory or anti-inflammatory roles in neuroinflammation caused by ischemic stroke, Parkinson's disease, and neuropathic pain. Lv et al. found that the expression of SNHG1 in HCMEC/D3 cells with oxygen-glucose deprivation condition was decreased compared with that in control cells. The luciferase reporter assay verified the targeting relationship between SNHG1 and miR-376a, and miR-376a had pro-inflammatory and pro-apoptotic effects. However, CBS/H2S can reverse the effects of miR-376a by reducing the release of IL-6, IL-1β, and TNF-α inflammatory factors in the OGD cell model and the transformation of microglia into M2 anti-inflammatory phenotype in the MCAO animal model (Zhang et al., 2017; Lv et al., 2021b). Meng et al. found that SNHG1 and NLRPS in C57BL/6 mice (intraperitoneal injection of MPTP-HCl) and BV2 cells (injection of LPS) were up-regulated. Moreover, SNHG1 bounded NLRP3 through competition with miR-7, which could promote microglial activation and inflammation and promote primary dopaminergic neurons apoptosis (Meng et al., 2021). CDKs are a family of Ser/Thr kinases. CDKs participate in physiologic and pathologic responses by regulating cell cycle. Importantly, CDKs can promote the expression of pro-inflammatory factors in G1 phase (Schmitz and Kracht, 2016). Zhang et al. found that SNHG1 bound to the CD4 promoter region and increased the release of inflammatory factors IL-6, TNF-α, IL-1β, and SNHG1 knockdown could alleviate neuropathic pain progression, suggesting that SNHG1 was a target for neuropathic pain clinical treatment (Zhang et al., 2020b).
Similar to the regulation of SNHG1 in neuropathic pain, SNHG4 overexpressed in model rats promoted neuroinflammation and neuropathic pain progression as a sponge molecule for miR-423-5P. Moreover, down-regulation of SNHG4 can reverse the increased expression of IL-6, IL-12, and TNF-α and the enhancement of mechanical allodynia and thermal hyperalgesia caused by depletion of miR-423-5p, suggesting that knockdown of SNHG4 may be a potential treatment of neuropathic pain and neuroinflammation (Pan et al., 2020). In the environment of cerebral ischemia and hypoxia, knockdown of AQP4 (the main water channel protein in the brain) reduced BBB damage and toxic edema to alleviate brain damage (Wang et al., 2015). Under the same conditions, TP53INP1 showed its pro-inflammatory and pro-apoptotic effects (Li et al., 2018e). In studies on ischemic stroke, both SNHG14 and SNHG15 formed miR-199b/AQP4 and miR-455-3p/TP53INP1 axis through the ceRNA network to promote glial cells and neurons apoptosis, oxidative stress, and inflammation. In oxygen-glucose deprivation-induced BV2 and PC12 cells, the levels of inflammatory factors (TNF-α and IL-1β), reactive oxygen species and malondialdehyde were up-regulated. In addition, knockdown of SNHG14 and SNHG15 could reverse the increased levels, suggesting that SNHG14 and SNHG15 were involved in the regulation of neuroinflammation and oxidative stress in ischemic stroke. Moreover, deletion of miR-199b and miR-455-3p could weaken the anti-inflammatory effects of SNHG14 and SNHG15 knockdown. However, the overexpression of downstream targets of miR-199b, and miR-455-3p attenuated the decreased expression of TNF-α and IL-1β (Fan et al., 2021; Zhang et al., 2021b). Different from their pro-inflammatory effects, the down-regulated expression of SNHG8 in primary microglial cells (treated with oxygen-glucose deprivation/reperfusion) was manifested as anti-inflammatory. SNHG8 could not only reverse the up-regulation of IL-1β, IL-6, and TNF-α in microglia caused by middle cerebral artery occlusion but also inhibit the release of pro-inflammatory factors in microglia caused by miR-425-5p. Mechanismally, SNHG8 was identified as the ceRNA of miR-425-5p, and the highly expressed SIRT1 in the model could be inhibited by miR-425-5p. SIRT1 regulates the oxidative respiration and cellular survival of hypothalamic neurons and can also deacetylation p65 to make NFκB inactivation. Therefore, SNHG8 regulated miR-425-5p/SIRT1 to inactivate downstream NFκB and inhibit microglial inflammation and BBB damage (Kauppinen et al., 2013; Tian et al., 2021).
GAS5
SNHG2, known as growth arrest specific 5 (GAS5), is located at chromosome 1q25.1. GAS5 is commonly expressed in the ovary, thyroid, bone marrow, and other tissues. MiR-196a negatively regulates GAS5 to inhibit esophageal squamous cell carcinoma growth. In breast cancer MCF-7 and MDA-MB-231 cells, miR-21 interacts with GAS5 ( Zhang et al., 2013; Wang et al., 2018b). In a physiologic state, GAS5/miR-18a/connective tissue growth factor was involved in the adipogenic differentiation of mesenchymal stem cells (Li et al., 2018c). GAS5/miR-23a/ATG3 is involved in autophagy and cell viability of 293T cells (Li et al., 2018b). GAS5/NODAL signal participates in the self-renewal of human embryonic stem cells (Xu et al., 2016). GAS5/miR-21/FGF1 is involved in the proliferation and apoptosis of growth plate chondrocytes (Liu et al., 2018).
As a biomarker, GAS5 is related to the risk of gastric cancer, uterine cervical cancer, and prostate cancer and is also a potential marker of sepsis inflammation (Lin et al., 2019; Dong et al., 2020; Weng et al., 2020; Zhang et al., 2022b). Additionally, GAS5 associated with miR-21 and miR-140 as potential markers of allergic rhinitis (Song et al., 2021).
As far as current studies are concerned, GAS5 is up-regulated in neurosystemic diseases and plays a pro-inflammatory role in neuroinflammation. Xu et al. found that overexpression of GAS5 could promote LPS-treated microglial inflammation, while miR-223-3p had the opposite effect by inhibiting NLRP3 inflammasome. As a competitive RNA, GAS5 reduced the expression of miR-223-3p, leading to the activation of the NLRP3 inflammasome. Thus, GAS5/miR-223-3p/NLRP3 plays an important role in the occurrence and development of Parkinson’s disease (Xu et al., 2020). Spinal cord ischemia in vivo study found that the expression of GAS5 and MMP-7 was abnormally high, and the expression of GAS5 was positively correlated with the expression of MMP-7. Knockdown of both GAS5 and MMP-7 could cause low expression of cleaved caspase-3 and IL-1β, which means that GAS5 and MMP-7 were related to apoptosis and inflammation in spinal cord ischemia. Therefore, knocking down GAS5 can alleviate apoptosis and inflammation after spinal cord ischemia-reperfusion through the MMP-7/cleaved caspase-3 axis (Zhang et al., 2021d). Sun et al. showed that GAS5 inhibited the differentiation of microglia into protective M2 phenotype. However, loss of the M2 phenotype leads to demyelination, exacerbating the course of demyelinating diseases similar to multiple sclerosis. PRC2 is a polymer composed of EZH1, EZH2, embryonic ectoderm development, and other subunits (Laugesen et al., 2019). EZH2 subunit is the main catalytic subunit of PRC2 complex, which can bind not only upstream GAS5 but also downstream IRF4 in multiple sclerosis. IRF4 is an important transcription factor in M2 phenotypic differentiation of microglia. Therefore, there exists PRC2/IRF4 axis in downstream of GAS5 to regulate M2 phenotype differentiation. In addition, it is speculated that there may be an IL-4/M-CSF-PI3K-mTOR axis in upstream of GAS5 (Sun et al., 2017).
H19
H19 imprinted maternally expressed transcript (H19) is located at chromosome 11p15.5. It is more often expressed in the placenta than in other tissues. LncRNA PTCSC3 regulates H19 to inhibit triple-negative breast cancer cell proliferation (Wang et al., 2019b). Transcription regulator HIF-1α upregulates H19 level, and knockdown of H19 improves the progression of liver fibrosis (Wang et al., 2020b). DEAD-box helicase 43 regulates H19 through demethylation and then increases its expression, which is involved in chronic myeloid leukemia (Lin et al., 2018). In the physiologic state, H19 regulates the differentiation of stem cells. The regulation of differentiation is not limited to osteogenic, neural-like, and adipocyte differentiation of bone marrow mesenchymal stem cells (Huang et al., 2016; Farzi-Molan et al., 2018; Ma et al., 2020a). Additionally, H19 regulates the odontogenic differentiation of human dental pulp stem cells (Zeng et al., 2018; Zhong et al., 2020).
The relationship between H19 SNPs and hepatoblastoma susceptibility has also been demonstrated (Tan et al., 2021). It has also become a diagnostic marker for gastric cancer and a prognosis marker for esophageal squamous cell cancer, papillary thyroid cancer, and glioblastoma (Jiao et al., 2019; Li et al., 2019b; Schaalan et al., 2020; Liu et al., 2021e).
In existing studies, H19 was upregulated in neuroinflammation and played a pro-inflammatory role. Rezaei et al. found that H19 was associated with ischemic stroke susceptibility and could be used as a diagnostic biomarker for ischemic stroke. The molecular mechanism of H19 in ischemic stroke was revealed by Wang et al. Histone deacetylases functioned as nerve injury in brain injury models. Its inhibitors, sodium butyrate and vorinostat, could improve the disease outcome by inhibiting the activation of microglia. In vitro, H19 targeted HDAC1 to inhibit the polarization of microglial M2 phenotype and promote neuroinflammation (Jaworska et al., 2017; Wang et al., 2017a ; Rezaei et al., 2021). Gu et al. found that H19 expression was up-regulated in the spinal cord injury model compared with control mice. Up-regulation of H19 expression could promote apoptosis and inflammation of BV2 cells. With interaction through the complementary base pairing of miR-325-3p and H19, it was found that miR-325-3p was the direct target gene of H19. MiR-325-3p is a tumor suppressor, which had a neuroprotective effect under ischemic and hypoxic stress conditions. It showed an inhibitory effect on inflammation in diabetic nephropathy. NEUROD4 is a harmful factor in the process of spinal cord injury (Yang et al., 2018c), and the targeted regulation of mir-325-3p can inhibit its expression, reducing glial inflammation and oxidative stress. Therefore, H19, miR-325-3p, and NEUROD4 formed a ceRNA network to regulate the disease process in spinal cord injury (Gu et al., 2021). Janus kinase/signal transducer and activator of transcription is a known signal pathway, which is related to cell proliferation, inflammation, and other processes. In the status epilepticus model (a status epilepticus model that could be constructed by microinjecting kainic acid into the amygdala of Sprague–Dawley rats is a common model of temporal lobe epilepsy), H19 could activate this pathway, which promoted the activation of astrocytes and microglia and the release of pro-inflammatory cytokines, suggesting that H19 could be a potential target for the treatment of epilepsy (Han et al., 2018).
Maternally expressed 3
Maternally expressed 3(MEG3) is located at chromosome 14q32.2. MEG3 is commonly expressed in the placenta, adrenal, and brain. DNA methyltransferase family can target the promoter of MEG3 to perform its regulation, so ubiquitin like with PHD and ring finger domains 1, pRb, and miRNA can regulate MEG3 by regulating DNMTs (Kruer et al., 2016; Zhuo et al., 2016; Cui et al., 2018). Interestingly, miRNA can also directly bind to the 3′‐untranslated region of MEG3 to regulate it (Zhou et al., 2015b). Under physiologic conditions, it has regulatory effects on stem cell differentiation, including promoting neural differentiation of human embryonic stem cells and osteogenic differentiation of bone marrow mesenchymal stem cells, inhibiting chondrogenic differentiation of synovium-derived mesenchymal stem cells (Mo et al., 2015; You et al., 2019; Li et al., 2021). In addition, MEG3/miR-128/Girdin can protect vascular endothelial cells from senescence (Lan et al., 2019), and MEG3 can regulate the proliferation of human umbilical vein cells proliferation through PTBP3/p53 (Shihabudeen Haider Ali et al., 2019).
As a biomarker, the SNP of MEG3 is associated with gastric cancer and oral squamous cell carcinoma susceptibility (Hou et al., 2019; Kong et al., 2020). MEG3 is a diagnostic marker for hunner type interstitial cystitis and chronic hepatitis B (Chen et al., 2019a; Liu et al., 2020a). In additiona, MEG3 functions as a diagnostic and prognostic marker in pancreatic cancer, mammalian cancer, and other cancers (Li et al., 2017; Ma et al., 2018).
MEG3 plays a pro-inflammatory or anti-inflammatory role in the occurrence and development of inflammation. In pulpitis, chronic pulmonary disease, and atherosclerosis, MEG3 presents a pro-inflammatory effect (Yan et al., 2019; Song et al., 2020; Liu et al., 2021a), whereas in ulcerative colitis, ankylosing spondylitis, and rheumatoid arthritis, MEG3 has an anti-inflammatory effect (Li et al., 2019a; Li et al., 2020c; Wang et al., 2021b). Pro-inflammatory MEG3 activates inflammasome by negatively regulating miRNA, thus promoting the occurrence and development of neuroinflammation. The five inflammasome types are AIM2, NLRP1, NLRP3, NLRC4, and IPAF inflammasome. AIM2 and NLRP3 were involved in cerebral ischemia reperfusion and traumatic brain injury-induced neuroinflammation under the regulation of MEG3/miR-485 and MEG3/miR-7a-5p, respectively (Liang et al., 2020; Meng et al., 2021). Yi et al. reported that MEG3 was down-regulated in Alzheimer's disease animal (Sprague–Dawley rats were injected Aβ25‐35) and played a protective role in disease progression. Overexpression of MEG3 reduced the expression of p-PI3K and p-Akt. The inhibition of the PI3K/AKT pathway inhibited activation of astrocytes;inhibited the release of IL‐1β, IL‐6, and TNF‐α; inhibited oxidative stress injury; and improved the cognitive impairment of Alzheimer’s disease rats. It has been suggested that MEG3 may function as a therapeutic target for Alzheimer's disease (Yi et al., 2019).
XIST
X inactive specific transcript (XIST) is located at chromosome Xq13.2. XIST is commonly expressed in the thyroid, ovary, and endometrium. Transcription factor Yin Yang 1 binds directly to the Xist 5′ region to activate XIST, and miR-7 regulates the downstream miR-92b/Slug/ESA axis by negatively regulating XIST (Makhlouf et al., 2014; Li et al., 2020a). Notably, m(6)A RNA methylation of XIST is required for XIST to perform its transcriptional inhibitory function (Patil et al., 2016). Under physiologic conditions, Xist promoted the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells through miR-9-5p/ALPL (Zheng et al., 2020). XIST inhibits Th17 cell differentiation through miR-377-3p/ETS1 (Yao et al., 2022).
XIST is a potential diagnostic marker of triple-negative breast cancer, colorectal cancer, and acute myocardial infarction, and 53BP1 combined with XIST is a prognostic marker of BRCA1-like breast cancer ( Schouten et al., 2016; Yu et al., 2020a; Lan et al., 2021; Zheng et al., 2022).
XIST plays a pro-inflammatory role in epilepsy and neuropathic pain by acting as a ceRNA of miRNA. Zhang et al. observed the expression of XIST and NFAT5 in CTX-TNA2 astrocyte cell line treated by LPS was significantly increased, while the expression of miR-29c-3p was decreased. Inhibition of miR-29c-3p or up-regulation of NFAT5 could reverse the inhibit astrocyte activation and inflammatory-induced neuronal apoptosis by XIST (Zhang et al., 2021c). It is important to note that the inflammation regulation of NFAT5 depends on the disease state; whether it plays a protective or damaging role depends on different stages of epilepsy development (Yang et al., 2018d, 2019b). XIST participated in the development of neuropathic pain as ceRNA. It was reflected in decreased paw withdrawal threshold and latency and increased level of pro-inflammatory cytokines. Therefore, XIST was considered as a potential therapeutic target for neuropathic pain and neuroinflammation. It also functioned through XIST/miR-150/ZEB1 and XIST/miR-544/STAT3 axis (Jin et al., 2018b; Yan et al., 2018).
UCA1
Urothelial cancer associated 1 (UCA1), formerly registered as bladder cancer invasion-associated gene, is located at chromosome 19p13.12. It is highly expressed in the gall bladder, endometrium, and urinary bladder. As its name means, UCA1 is first found in abnormally high expression on bladder cancer tissue. Meanwhile, UCA1 is related to the progression of bladder cancer (Wang et al., 2006). The regulation of UCA1 can be realized by transcription factors CCAAT/enhancer-binding protein α, HIF-1α, and SP1 binding to the UCA1 promoter. Moreover, it can be regulated by insulin-like growth factor 2 messenger RNA binding protein. Even lncRNA GAS8-AS1 and hsa-miR-1 can negatively regulate UCA1 expression (Wang et al., 2014, 2017b; Jin et al., 2018a; Zhou et al., 2018c; Li et al., 2018d; Zha et al., 2020). UCA1 plays an important regulatory role in physiologic and pathologic processes. In a physiologic state, UCA1 regulates cell proliferation, differentiation, migration, and epithelial-mesenchymal transition (Ishikawa et al., 2018; Liu et al., 2020b; Yu et al., 2020c; Zhang et al., 2020c).
As a biomarker, it is mainly involved in screening, diagnosis, and prognosis and is related to cancer and inflammation. In combination with H19, it is associated with the susceptibility to 5-fluorouracil in rectal cancer (Yokoyama et al., 2019). Combined with PGM5-AS1, it becomes the diagnostic marker of early-stage colorectal cancer (Wang et al., 2021a), and it is related to the multiple pro-inflammatory cytokines of sepsis patients and acute stroke patients (Ren et al., 2021; Wang et al., 2022). Furthermore, combined with NEAT1, it predicts poor prognosis in oral squamous cell carcinoma (Zhu et al., 2021). As an oncogenic gene, it regulates the proliferation, apoptosis, epithelial-mesenchymal transition, invasion, metastasis, and chemoresistance of gastric cancer cell lines, colon cancer cells, breast cancer cells, and other cancer cells (Cao et al., 2020; Yang et al., 2021; Wo et al., 2022).
The regulatory role of UCA1 in inflammation has been clarified in recent years, and its expression and inflammatory effects have ambivalent effects on different diseases. Yu et al. reported that UCA1 was underexpressed in an epilepsy model in vitro and in vivo. Further studies revealed that UCA1, as a ceRNA, promoted the expression of MEF2C by adsorbing miR-203. Notably, MEF2C was able to not only inhibit IKK expression and IkBα phosphorylation but also inhibit phosphorylation of p65 Ser 536 and p65 nuclear translocation. In addition, the UCA1/miR-203/MEF2C/NF-κB pathway could improve IL-1β-treated CTX-TNA2 cells apoptosis and inhibit the expression of IL-6, TNF-α and COX-2 (Xu et al., 2015; Yu et al., 2020b). In contrast, in the Parkinson’s disease model with injection of 6-OHDA of Wistar rats, the overexpression of UCA1 showed a pro-inflammatory effect. UCA1 promoted the expression of p-PI3K and p-AKT as well as the phosphorylation of IKBα and ERK, which were downstream molecules of the PI3K/AKT pathway. Transfection of siRNA-UCA1 up-regulated the expression of nerve growth factor brain-derived neurotrophic factor and decreased the levels of malondialdehyde, TNF-α, IL-6, and IL-1β. It is suggested that UCA1 may play an essential role in dopaminergic neurons apoptosis, oxidative stress, and inflammation in Parkinson’s disease (Cai et al., 2019). A study has found that the expression of UCA1 was related to the expressions of cytokines including TNF-α, IL-6, and IL-17 in sepsis. In another study, UCA1/EZH2/HOXA1 was found to play a regulatory role in sepsis-induced pneumonia (Wang et al., 2022; Zhang et al., 2022c). The expression of UCA1 is positively correlated with the cell ratio of Th17, IL-17, and IL-6 in acute ischemic stroke patients. It is speculated that UCA1 is related to the inflammatory response in the pathologic process of acute ischemic stroke, and the related mechanism needs to be further studied (Ren et al., 2021).
GM4419
Predicted gene 4419 (Gm4419) is located at chromosome 12A1.3. The GM4419 expression of testis adult is higher than other tissues. Currently, there are few studies on GM4419.
Existing studies have shown that GM4419 was abnormally overexpressed in diabetic nephropathy, ischemic stroke, and traumatic brain injury and played a pro-inflammatory role in the disease process. Gm4419 not only binded to IκBα to promote the phosphorylation of IκBα but also directly interacted with p50, thereby promoting nuclear translocation of p65/p50. Furthermore, the release of cytokine IL-6, TNF-α, and IL-1β promoted the inflammation of oxygen-glucose deprivation/reperfusion-treated microglial cells. Yi et al. showed that the NFκB pathway activated by GM4419 could trigger the activation of NLRP3 inflammasome and promote the process of diabetic nephropathy (Wen et al., 2017; Yi et al., 2017). Yu et al. found that Gm4419 directly inhibited miR-466l, leading to up-regulation of TNF-α in trauma-induced astrocyte, which was related to astrocyte apoptosis. These results suggested that GM4419, miR-466l, TNF-α, and astrocyte apoptosis were involved in the process of traumatic brain injury (Yu et al., 2017).
Clinical Significance
The clinical significance of lncRNA is that it can be used as a biomarker for diagnosis and prognosis of disease and as a target for disease treatment. Individual lncRNA or complexes formed in combination with other miRNA may be effective biomarkers of disease.
With the in-depth study of the molecular mechanism of lncRNA production in the context of disease, lncRNA-targeted therapeutic are promising. At present, lncRNA-targeted drug technologies involve small molecules and gene therapies. The mode of small molecules is to structurally bind lncRNA specifically, destroy the spatial structure of target lncRNA, and regulate the level of endogenous lncRNA. Studies have shown that NP-C86 small molecule and Rod-like DPFs ligand can respectively act on lncRNA GAS5 and lncRNA MALAT1 (Donlic et al., 2018; Shi et al., 2019b). Gene therapies based on clustered regularly interspaced short palindrome repeats technology, antisense RNA technology, and RNAi technology have their own unique modes. The former system relies on sequence recognition of single guide RNA and sequence cleavage of Cas9 enzyme, which can act on both nuclear and cytosolic lncRNAs. Antisense RNA technology involves targeting lncRNAs to activate RNase H, which degrades target lncRNAs. The antisense oligonucleotide drugs Nusinersen, Eteplirsen, and Inotersen have been applied in spinal muscular atrophy, Duchenne muscular dystrophy, and familial amyloid polyneuropathy, respectively. RNAi is about silencing genes by siRNA. The siRNA drugs Onpattro and Givlaari have been used in polyneuropathy and acute hepatic porphyrin. Compared with gene therapies, small molecule drugs are cheaper and easier to administer but less selective. In contrast, gene therapy can be largely programmed and personalized medicine. Currently, researchers are also optimizing the combination of drug technologies and drug delivery systems.
Conclusion and Perspective
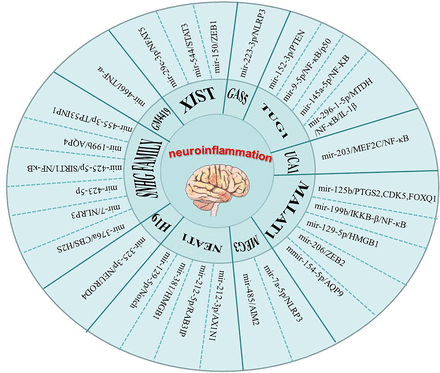
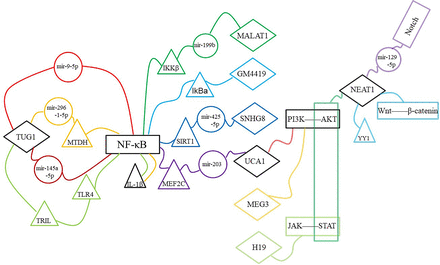
Among the 10 lncRNAs introduced in this paper, ceRNA network is the main regulatory mechanism of neuroinflammation, (Fig. 2) and NFκB pathway is the main inflammatory pathway involved in neuroinflammation (Fig. 3). There is a feedback mechanism between pathologic processes. On the one hand, neuroinflammation may be triggered by molecules in other pathologic processes, and, on the other hand, neuroinflammation may induce other pathologic processes. Accumulational studies on lncRNAs have shown that lncRNAs exert non-single effects on cell functions under physiologic and pathologic conditions. Therefore, targeting lncRNAs related to inflammation can not only alleviate cell damage caused by neuroinflammation but also improve brain function of individuals with disease. At the same time, we realize that a single drug cannot deal with multiple pathologic processes in the disease process, so exosomes containing multiple components in combination with existing drug development technologies may be a good therapeutic option.
CeRNA networks in neuroinflammation. Among the mechanisms by which lncRNAs regulate neuroinflammation, the ceRNA network has been studied more widely. CeRNA network consists of lncRNA, miRNA, and mRNA. Specifically, lncRNA targets the 3′-UTR of miRNA to regulate downstream mRNA.
Signal pathways in neuroinflammation. Neuroinflammation-related signaling pathways include NFκB, Notch, PI3K/AKT, and so on. NFκB is the main pathway of lncRNAs to regulate neuroinflammation.
Author Contributions
Wrote or contributed to the writing of the manuscript: Zeng, Zhang, Lin, Lin, Wu.
Acknowledgments
We have extensively searched several databases, PubMed, and Web of Science to collect the information, the articles and the reference lists were examined in depth.
Footnotes
- Received May 20, 2022.
- Accepted November 7, 2022.
This work was supported by the National Natural Science Foundation of China [Grant 81801189].
No author has an actual or perceived conflict of interest with the contents of this article.
An earlier version of this paper appears in Neuroinflammation in Neurologic Diseases: The Roles of LncRNAs under the doi MOLPHARM-PI-2022-000526-T.
Abbreviations
- AIM2
- absent in melanoma 2
- BBB
- blood-brain barrier
- BDNF
- brain-derived neurotrophic factor
- CDK5
- cyclin dependent kinase 5
- ceRNA
- competing endogenous RNA
- C/EBPβ
- CCAAT/enhancer-binding protein Β
- CNR2
- cannabinoid receptor 2
- COX-2
- cyclooxygenase-2
- CPSF6
- cleavage and polyadenylation specific factor 6
- CREB
- camp response-element binding protein
- CRISPR/Cas9
- clustered regularly interspaced short palindrome repeats
- CTGF
- connective tissue growth factor
- DANCR
- differentiation antagonizing non-protein coding RNA
- DDX43
- DEAD-box helicase 43
- DOT1L
- disruptor of telomeric silencing 1-like protein
- EED
- embryonic ectoderm development
- EZH2
- enhancer of zeste 2 polycomb repressive complex 2 subunit
- FGF1
- fibroblast growth factor 1
- FOXQ1
- forkhead box Q1
- GAS5
- growth arrest specific 5
- Gm4419
- predicted gene 4419
- H19
- H19 imprinted maternally expressed transcript
- hASC
- human adipose-derived stem cell
- HES1
- Hes family Bhlh transcription factor 1
- HIF-1α
- hypoxia-inducible factor-1Α
- HMGB1
- high mobility group box protein 1
- HUVECs
- human umbilical vein endothelial cells
- IL-6/1β/17
- interleukin-6/1Β/17
- JAG1
- jagged canonical notch ligand 1
- JAK/STAT
- Janus kinase/signal transducer and activator of transcription
- KIF4
- Kruppel like factor 4
- lncRNA
- long noncoding RNA
- LPS/ATP
- lipopolysaccharide/adenosine triphosphate
- M6A and M5C
- N6-methyladenosine and 5-methylcytosine
- MALAT1
- metastasis-associated lung adenocarcinoma transcript 1
- MCAO
- middle cerebral artery occlusion
- MEF2C
- myocyte enhancer factor 2C
- MEG3
- maternally expressed 3
- miRNA
- micro RNA
- MMP-7
- matrix metallopeptidase 7
- mTOR
- mechanistic target of rapamycin
- NEAT1
- nuclear paraspeckle assembly transcript 1
- NEUROD4
- neuronal differentiation 4
- NFAT5
- nuclear factor of activated T cells 5
- NF-κB
- nuclear factor kappa light chain enhancer of activated B cells
- NLRC4
- NLR family CARD domain containing 4 protein
- NLRP3
- Nlr family pyrin domain containing 3
- NRF2
- nuclear factor erythroid 2-related factor 2
- OGD/R
- oxygen-glucose deprivation/reperfusion
- PI3K/AKT
- phosphatidylinositol-3-kinase/Ak strain transforming
- PTBP3
- polypyrimidine tract binding protein 3
- PIDD1
- P53-induced death domain protein 1
- PPAR
- peroxisome proliferator-activated receptor
- PRC2
- polycomb repressive complex 2
- PTEN
- phosphatase and tensin homolog
- PTGS2
- prostaglandin-endoperoxide synthase 2
- RNAi
- RNA interference
- SK1
- sphingosine kinase 1
- SNHG1/8/14/15
- small nucleolar RNA host gene 1/8/14/15
- SNP
- single nucleotide polymorphism
- SR
- serine/arginine-rich family splicing factor
- SRSF1
- serine and arginine rich splicing factor 1
- TIMP2
- TIMP metallopeptidase inhibitor 2
- TNF-α
- tumor necrosis factor alpha
- TP53INP1
- tumor protein P53 inducible nuclear protein 1
- TUG1
- taurine upregulated 1
- UCA1
- urothelial cancer associated 1
- UTR
- untranslated regions
- VEGFA
- vascular endothelial growth factor A
- XIST
- X inactive specific transcript
- YAP1
- yes-associated protein 1
- ZEB2
- zinc finger E-box binding homeobox 2
- Copyright © 2023 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.