Abstract
Histamine is a multifunctional hormone that regulates smooth muscle contraction in the airways, acid secretion in the gut, and neurotransmitter release in the central nervous system through three well characterized receptor subtypes, H1, H2, H3, respectively. As part of a directed effort to discover novel G-protein-coupled receptors through homology searching of genomic databases, we identified a partial clone (GPCR105) that had significant homology to the recently identified histamine H3 receptor cDNA. Expression of the full-length human GPCR105 in cells confers the ability to bind [3H]histamine with high affinity (K D = 5 nM). GPCR105 is pharmacologically similar to the histamine H3 receptor in that it binds many of the known H3 agonists and antagonists, albeit with a different rank order of affinity/potency. GPCR105 does not bind (i.e., K D > 10 μM) all tested H1 and H2 receptor antagonists such as diphenhydramine, loratadine, ranitidine, and cimetidine, but has modest affinity for the H2 receptor agonist, dimaprit (377 nM). Whereas the H3 receptor is expressed almost exclusively in nervous tissues, GPRC105 is expressed primarily in bone marrow and eosinophils. Together, these data demonstrate that GPCR105 is a novel histamine receptor structurally and pharmacologically related to the H3 receptor. However, its unique expression profile and physiological role suggest that GPCR105 is a fourth histamine receptor subtype (H4) and may be a therapeutic target for the regulation of immune function, particularly with respect to allergy and asthma.
Histamine was first identified as a hormone in 1910 (Barger and Dale, 1910) and has since been demonstrated to play a major role in a variety of physiological processes, including the inflammatory “triple response” via H1 receptors (Ash and Schild, 1966), gastric acid secretion via H2 receptors (Black et al., 1972), and neurotransmitter release in the central nervous system via H3 receptors (Arrang et al., 1983; for a review see Hill et al., 1997). All three histamine receptor subtypes have been demonstrated to be members of the superfamily of G-protein-coupled receptors (Gantz et al., 1991; Yamashita et al., 1991; Lovenberg et al., 1999). There are, however, additional functions of histamine that have been reported, for which no receptor has been identified. For example, in 1994, Raible et al. (1994) demonstrated that histamine and (R)-α-methylhistamine could activate calcium mobilization in human eosinophils. These responses were blocked by the H3-receptor antagonist thioperamide. However, (R)-α-methylhistamine was significantly less potent than histamine, which was not consistent with the involvement of known H3 receptor subtypes. Therefore, Raible et al. (1994) hypothesized the existence of a novel histamine receptor on eosinophils that was non-H1, -H2, -H3.
In an effort to search for additional members of the G-protein-coupled receptor family, we performed homology searching of the human genome databases using the sequences of all known receptors as queries. This has led us to the identification of over 100 putative novel receptors, one of which (GPCR105), was highly related in sequence to the histamine H3 receptor. GPCR105 was identified in a series of unordered fragments from chromosome 18 (GenBank accession no.AC007922) and when assembled in an appropriate orientation exhibits an exon/intron arrangement nearly identical to that of the H3 receptor gene. Expression of the full-length human cDNA of GPCR105 in cells confers the ability to bind histamine. We therefore investigated the possibility that GPCR105 represented the novel histamine receptor proposed by Raible et al. (1994).
Experimental Procedures
Materials
Human mRNA was purchased from CLONTECH (Palo Alto, CA). cDNA synthesis kits were purchased from Life Technologies, Inc. (Gaithersburg, MD). Gelzyme was obtained from Invitrogen (San Diego, CA) and pCIneo vector was purchased from Promega (Madison, WI). All cell lines were obtained from American Type Culture Collection (American Type Culture Collection, Manassas, VA). cAMP Flashplates were from PerkinElmer Life Sciences (Boston, MA). G418 was purchased from Calbiochem (San Diego, CA). All histamine receptor ligands were either purchased from RBI (Natick, MA) or synthesized in-house. Burimamide was synthesized according to the method of Vollinga et al. (1995). 4-(3-Piperidin-1-yl-propoxy)-benzonitrile was synthesized according to the method of Schwartz et al. (1998). All other reagents were purchased from Sigma (St. Louis, MO).
Cloning Human GPCR105 cDNA
The complete coding region of human G-protein-coupled receptor (GPCR) cDNA was PCR amplified using two primers P1: 5′-ACT AGA ATT CGC CAC CAT GCC AGA TAC TAA TAG CAC A-3′; and P2: 5′-ACT ACT GCG GCC GCT TAA GAA GAT ACT GAC CGA CTG T-3′ with human spleen cDNA (CLONTECH) as the template. The cDNA was cloned into expression vector pCINeo (Promega) between the EcoRI and NotI site. The entire insert was sequenced using an automated DNA sequencer (Prizm 377, PerkinElmer). The complete cDNA sequence of GPCR105 including the 5′-untranslated region (UTR) and the 3′-untranslated region was obtained by a combination of screening a human bone marrow cDNA library and 5′-rapid amplification of cDNA ends (RACE) using human bone marrow marathon cDNA (CLONTECH) as template. To RACE the 5′-untranslated region, human GPCR105 specific primer P3: 5′-ACG TGA ATT CTA TAT ACA GAT GCT GTA CAT AAC-3′ and adaptor primer AP1: 5′-CCA TCC TAA TAC GAC TCA CTA TAG GGC-3′ (CLONTECH) were used. The PCR was performed as described by the manufacturer. The final RACE PCR product was then cloned into a plasmid pBluescript (Stratagene, La Jolla, CA) and the insert sequenced. The 3′-end-untranslated region was obtained by screening a human bone marrow cDNA library using a cDNA probe containing the entire GPCR105 coding region.
Cell Cultures
Human GPCR105 or human H3 receptor full-coding region in pCINeo were transfected into SK-N-MC cells using the LipofectAMINE method (Life Technologies, Inc.) according to the manufacturer's instructions. A cell line stably expressing GPCR105 was established in SK-N-MC cells by selection in the presence of G418 (400 mg/liter). HL-60 clone 15 cells were obtained from the ATCC (Manassas, VA) and maintained in RPMI 1640 with 10% fetal bovine serum,l-glutamine, and antibiotics. Eosinophilic differentiation was performed as described by Tiffany et al. (1998). Briefly, cells were cultured in the presence of 0.5 mM butyric acid for 48 h. IL-5 was added to 10 ng/ml, and the cells were further cultured for 72 h. Eosinophilic maturation was determined by fluorescence-activated cell sorter analysis of anti-CCR3 stained cells.
GPCR105 mRNA Tissue Expression Pattern
Reverse Transcription-PCR Detection.
Using primers P1 above and P4: 5′-ATG CAG GAT CCA GCA TTT GAG ACT GAC AGG TAT-3′, we have analyzed multiple tissue cDNAs under condition of 94°C 45 sec, 60°C 45 sec, 72°C 2 min for 35 cycles. The PCR products were run on a 1% agarose gel, and DNA was stained with ethidium bromide (10 ug/ml) and visualized with UV. The PCR products in gel were then transferred to a nitrocellulose membrane and hybridized with a32P-labeled human GPCR105 DNA probe.
RNase Protection Detection.
A 350-bp fragment of human GPCR105 coding region was subcloned into the plasmid pBluescript, and the insert region was sequenced. 32P-Labeled antisense ribo-probe was generated from the linearized plasmid using T7 RNA polymerase. Total RNA (20 μg) from different tissues or cells were used to hybridize with the GPCR105 antisense as described bySambrook et al. (1989).
Northern Blot.
Human eosinophils were isolated from the buffy coat of 500 ml of blood anticoagulated with sodium citrate. Red blood cells were lysed, and mononuclear cells were centrifuged over isotonic Percoll. Neutrophils were removed by magnetic depletion using Dynabeads. The purity of eosinophils was >97%. HL-60 clone 15 cells (ATCC) were stimulated for 5 days with 0.5 mM butyric acid and IL-5 (see above). From unstimulated and stimulated HL-60 clone 15 cells and SK-N-MC cells expressing GPCR105, RNA was prepared using a RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA samples (5 μg) were run on an RNA gel and then transferred overnight to a nylon blot (Hybond, Amersham Pharmacia Biotech, Piscataway, NJ). The blot was prehybridized with ExpressHyb solution (CLONTECH) for 30 min at 68°C. The human GPCR105 vector insert was labeled using the rediprime II kit (Amersham Pharmacia Biotech). The blot was hybridized for 2 h at 68°C, followed by one wash step (2× SSC and 0.05% SDS) of 40 min at room temperature, and a second wash step (0.1× SSC and 0.1% SDS) of 40 min at 50°C. The blot was exposed to X-ray film at −70°C with two intensifying screens overnight.
Cyclic AMP Accumulation
A subline of SK-N-MC cells was created that expressed both GPCR105 and a reporter gene construct. The reporter gene was β-galactosidase under the control of cyclic AMP responsive elements. Cells were plated in 96-well plates the night before the assay. To start the assay, agonists were added directly to the cell medium followed 10 min later by an addition of forskolin (10 μM final concentration). Cells were returned to the incubator for 6 h at 37°C. The medium was then aspirated, and the cells were washed with 200 μl of PBS followed by a second aspiration. Cells were lysed with 25 μl of 0.1× assay buffer (10 mM sodium phosphate, pH 8, 0.2 mM MgSO4, 0.01 mM MnCl2) and incubated at room temperature for 10 min. Cells were then incubated for 10 min with 100 μl of 1× assay buffer containing 0.5% Triton and 40 mM betamercaptoethanol. Color was developed using 25 μl of 1 mg/ml substrate solution (chlorophenol red β-d-galactopyranoside; Roche Molecular Biochemicals, Indianapolis, IN). Color was quantitated on a microplate reader at absorbance 570 nm.
[3H]Histamine Binding
Cell pellets from GPCR105-expressing SK-N-MC cells were homogenized in 20 mM Tris-HCl/0.5 mM EDTA. Supernatants from an 800g spin were collected and recentrifuged at 30,000g for 30 min. Pellets were rehomogenized in 50 mM Tris/5 mM EDTA. Membranes were incubated with 5 nM [3H]histamine with or without test compounds for 45 min at 25°C. Nonspecific binding was defined with 10 μM cold histamine. In some experiments, [3H](R)-α-methylhistamine or [3H]N α-methylhistamine was used as the radioligand, but all other conditions were identical. For competition binding studies using [3H]histamine, K Ivalues were calculated based on an experimentally determinedK D value of 5 nM and a ligand concentration of 5 nM according to Cheng and Prusoff (1973).
Results
Cloning and Sequence Analysis of Human GPCR105.
A human genomic database BLAST search (Altschul et al., 1990) using the histamine H3 receptor as a query sequence revealed an unordered draft contiguous sequence (accession no.AC007922) encoding a putative novel GPCR, designated GPCR105. The complete coding region was predicted using the Genewise program (The Sanger Center). Putative intron/exon junctions were in identical regions as those predicted for the H3 receptor (Fig. 1). Primers were designed based upon the predicted start and stop codons, and the full-length clone was PCR-amplified from human spleen cDNA. The cDNA sequence indicated a single open reading frame encoding 390 amino acids. Using a 5′-RACE method, 100 base pairs of 5′-untranslated region were cloned. DNA sequence of the 5′-UTR shows that three in-frame stop codons are found beyond the predicted translation initiation methionine, suggesting that the proper start codon was identified. The 3′-UTR of human GPCR105 is 2415 base pairs and was isolated from a human bone marrow cDNA library using the entire coding region as a probe. The amino-acid sequence of GPCR105 is approximately 35% identical to the histamine H3 receptor, whereas it is less than 25% identical to all other receptors, including H1and H2 histamine receptors.
Protein amino acid sequence comparison between human GPCR105 and histamine H3 receptor. The putative seven-transmembrane regions were denoted in bold letters and underlined. The positions of introns are indicated by ↑. Nucleic acid sequence and amino acid sequence of GPCR105 have been deposited with GenBank (accession no. AF312230).
Tissue Expression Pattern of Human GPCR105 mRNA.
A preliminary PCR screen of several human cDNA libraries indicated abundant expression of GPCR105 in bone marrow, with little expression observed in other cDNA libraries (Fig. 2). To obtain a more quantitative assessment of expression, we designed an RNase protection assay (RPA). Using RPA, we analyzed GPCR105 expression in mRNA isolated from 15 different human tissues. The results demonstrate that the highest level of human GPCR105 expression is seen in human bone marrow and spleen. However, trace amounts of mRNA expression were detected in many different tissues (Fig.3). Expression of GPCR105 was evaluated by Northern blot in human eosinophils, and the promyelocytic cell line HL-60 clone 15 (CRL 1964; ATCC). Human eosinophils showed significant expression of GPCR105 (Fig. 4). In addition, the HL-60 clone 15 cells showed little endogenous expression of GPCR105, but expression could be clearly detected in the HL-60 clone 15 cells that had been differentiated to the eosinophilic phenotype with butyric acid/IL-5 for five days (Fig. 4). Expression was also clearly detected in the GPCR105-transfected SK-N-MC cells, whereas no expression was seen in untransfected, human H2 or H3 receptor transfected cells.
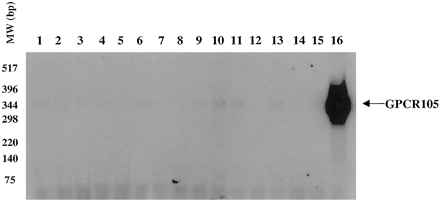
Detection of GPCR105 mRNA expression in different human tissues by reverse transcription-PCR. cDNAs from different human tissues (lanes 1 to 16) were used as templates for PCR amplification using human GPCR105 primers P1 and P4. Lanes: 1, cerebellum; 2, cortex; 3, hypothalamus; 4, small intestine; 5, dorsal root ganglia; 6, hippocampus; 7, spleen; 8, thalamus; 9, placenta; 10, heart; 11, liver; 12, lung; 13, uterus; 14, pituitary; 15, spinal cord; 16, bone marrow. The PCR products were run on a 1% agarose gel, transferred onto a nitrocellulose membrane, and blotted with a32P-labeled human GPCR105 DNA probe.
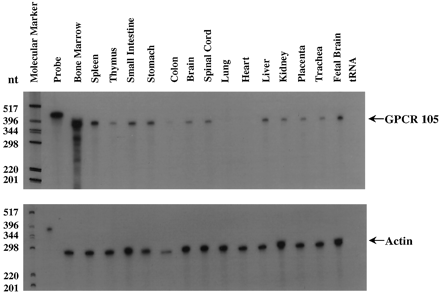
Detection of GPCR105 mRNA expression in different human tissues by RPAs. Twenty micrograms of total RNA each from different human tissue were used in RPA. As internal controls, RPA for human β-actin was performed in parallel. The RPA products were run on a 5% acrylamide sequencing gel. The gel was dried and exposed to a X-ray film overnight. nt, nucleotide.
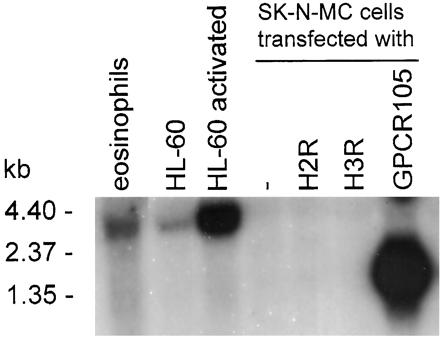
Human eosinophils and eosinophil-like cells express GPCR105. Northern blots of human eosinophils and human HL-60 clone 15 cells before and after 5 days of stimulation with butyric acid and IL-5. RNA from SK-N-MC cells stably transfected with GPCR105 was used as a positive control, while untransfected, H2 and H3 receptor transfected cells were used as a negative control. The predicted size of the GPCR105 mRNA (in tissue) is approximately 3700 bp, and the predicted size of the recombinant GPCR105 in SK-N-MC cells is 1300 bp. The difference in size is due to the natural receptor mRNA having 2400 bp of 3′-untranslated region, whereas this region is deleted in the recombinant construct. kb, kilobase(s).
GPCR105 Binds [3H]Histamine.
Because the protein sequence of GPCR105 is closest to human histamine H3 receptor (greater than 50% homology in the putative transmembrane domains), we tested the binding of several selective H3 radioligands. GPCR105 was transfected into SK-N-MC cells, and individual colonies surviving G418 selection were tested for binding with the H3selective radioligands [3H](R)-α-methylhistamine (0.8 nM), [3H]N α-methylhistamine (0.8 nM), and [3H]histamine (10 nM). These concentrations of radioligand were chosen because they match theirK D values for binding to the H3 receptor (Lovenberg et al., 1999; and unpublished observations). Surprisingly, binding of [3H](R)-α-methylhistamine and [3H]N α-methylhistamine to GPCR105-transfected cells was nearly undetectable at the tested concentrations. However, specific binding of 10 nM [3H]histamine was clearly detected. Subsequent saturation studies revealed that these three radioligands bind GPCR105, albeit with different K D values than for the H3 receptor (Table1). [3H]Histamine was the highest affinity ligand for GPCR105, whereas [3H](R)-α-methylhistamine was the highest for the H3 receptor. A representative saturation isotherm for [3H]histamine binding to GPCR105 is shown in Fig. 5. Subsequent competition binding experiments were performed with 5 nM (i.e.,K D) [3H]histamine. A variety of known H1, H2and H3 receptor ligands were evaluated for binding to GPCR105. GPCR105 displays modest affinity for histamine and a number of the known imidazole-based H3 ligands including the H3 agonists immepip, imetit,N-methylhistamine, (R)-α-methylhistamine, and the H3 antagonists clobenpropit, thioperamide, and burimamide (Table 2). The rank order of potency is noticeably different between the two receptors. [3H]Histamine binding to GPCR105 was unaffected (i.e., K I < 10 μM) by the H1 antagonists diphenhydramine and cyproheptadine, or the H2 antagonists ranitidine and cimetidine (Table 2). Like the H3 receptor, GPCR105 displayed modest affinity for the H2agonists dimaprit and impromidine. In fact, those compounds, along with clozapine, are the only ligands with higher affinity for GPCR105 than the H3 receptor. We also synthesized and tested several non-imidazole H3 antagonists that have recently been reported in the literature and patent literature (Ganellin et al., 1998; Schwartz et al., 1998). One of these compounds, 4-(3-piperidin-1-yl-propoxy)-benzonitrile, was tested at both the H3 receptor and GPCR105. 4-(3-piperidin-1-yl-propoxy)-benzonitrile had high affinity for the H3 receptor (K i = 25 nM), which is consistent with the cited value of 11 nM (Schwartz et al., 1998). However, it did not bind to GPCR105.
The affinity of radioligands for GPCR105 and the human H3receptor
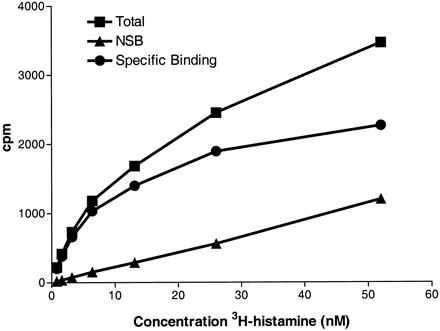
[3H]Histamine binding to GPCR105-transfected cells. Membranes from SK-N-MC cells stably transfected with GPCR105 were incubated with increasing concentrations of [3H]histamine. Nonspecific binding (NSB) was determined in the presence of 10 μM histamine. Linear regression of the line produced by a bound versus bound/free resulted in aK d of approximately 5 nM.
Affinity values (n = 3, average ± S.D.) of histamine ligands for the human H3 receptor on GPCR105
GPCR105 Is Weakly Coupled to Inhibition of Adenylate Cyclase.
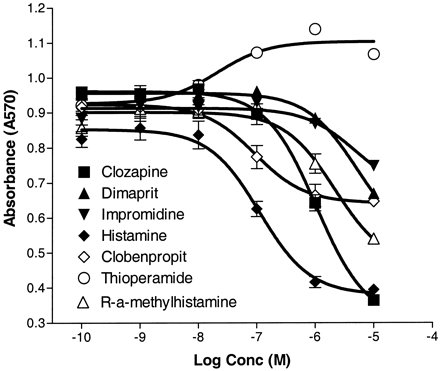
We have previously demonstrated that the H3receptor mediates inhibition of forskolin-stimulated adenylate cyclase when transfected into SK-N-MC cells (Lovenberg et al., 1999). GPCR105-transfected SK-N-MC cells also respond to histamine via inhibition of adenylate cyclase as well. However, the maximal inhibition achieved in the GPCR105-transfected cells was significantly less than that obtained with histamine in H3-transfected cells and thus made absolute quantification of the cAMP levels problematic. Therefore to obtain a reliable and measurable response, we transfected GPCR105 into a cell line expressing a cAMP-responsive element linked to a reporter gene output. Forskolin-stimulated reporter gene activity could be inhibited by histamine (Fig. 6). The corresponding EC50 value (approximately 50 nM) was 10-fold higher than the K D for [3H]histamine measured in the radioligand binding assay of GPCR105. In separate experiments, histamine did not stimulate basal cAMP levels or stimulate calcium mobilization in GPCR105-transfected SK-N-MC cells, nor did it affect forskolin-stimulated reporter gene activity in untransfected cells (not shown). We also tested impromidine, dimaprit, clobenpropit, thioperamide, clozapine, and (R)-α-methylhistamine. Interestingly, clobenpropit behaved as a partial agonist. Dimaprit and impromidine both showed some agonist activity up to 10 μM. Clozapine behaved as a full agonist in the assay (Fig. 6). All EC50 values were higher than the correspondingK I values obtained in the binding assays. Transfection of GPCR105 into COS7, HEK293, or CHOK1 cells did not improve its functional coupling (not shown). It remains to be determined whether coexpression of GPCR105 with various G-protein subunits can alter the efficiency of the effector coupling.
Inhibition of forskolin-stimulated cAMP accumulation by histamine and various ligands. Cyclic AMP accumulation was measured indirectly using a cAMP-sensitive reporter gene where the readout was β-galactosidase activity at 570 nm. Shown are the mean and S.E.M. of triplicate data points. This graph is representative of at least three similar experiments.
Discussion
Histamine plays a role in numerous physiological processes including gastrointestinal function, CNS function, and immune cell function (Hill et al., 1997; Bertaccini et al., 2000; Coruzzi et al., 2000). Effects of H1 and H2receptor ligands have been demonstrated on monocytes, eosinophils, mast cells, neutrophils, etc. (Tedeschi et al., 1991; Ley, 1994; Foster and Cunningham, 1998), although relatively little literature exists documenting H3-like effects on bone marrow-derived cells (Bissonnette, 1996). However, working with human eosinophils, Raible et al. (1994) demonstrated that histamine could dose dependently stimulate calcium mobilization in a thioperamide-sensitive manner. This effect could be mimicked, albeit with lower affinity, by N-methylhistamine, and (R)-α-methylhistamine. Dimaprit, the H2 agonist/weak H3antagonist also mimicked the effect of histamine. In a separate report,Schwoerer et al. (1994) demonstrated a histamine H3-like effect on 5-hydroxytryptamine release from porcine enterochromaffin-like cells.
Our current study identifies an orphan receptor, GPCR105, that exhibits properties consistent with a novel H3-like receptor. GPCR105 is most similar in amino acid sequence to the histamine H3 receptor (greater than 35% identity) as compared to all other G-protein- coupled receptors (25% identity). The putative intron/exon structure of GPCR105 is nearly identical to that predicted for the H3 receptor. Together, these data suggest a close relationship between these two receptors. When transfected into cells, GPCR105 binds [3H]histamine with high affinity in a saturable manner. Interestingly, GPCR105 also binds the “selective” H3 ligands (R)-α-methylhistamine andN-methylhistamine, but with low affinity relative to the H3 receptor. In fact, GPCR105 bound almost all known H3 ligands but with a unique affinity order. Thus, most of the H3 ligands expressed approximately 10-fold lower affinity for GPCR105 with the exception of (R)-α-methylhistamine, with 200-fold lower affinity for GPCR105. There were only three compounds (impromidine, clozapine, and dimaprit) that showed higher affinity at GPCR105 than at the H3 receptor. Interestingly, two of these compounds, impromidine and dimaprit, are better known for their H2 receptor properties, both of which are H2 agonists. All three of these compounds behaved as agonists (impromidine and dimaprit were partial agonists). Clozapine appeared to be a full agonist and was in fact more potent than (R)-α-methylhistamine. The H3antagonist clobenpropit surprisingly behaved as a partial agonist in the assay. Thioperamide showed what would appear to be inverse agonism in that it enhanced cAMP activity. Understanding this complicated pharmacology will be critical to understanding the true physiological role of the GPCR105 receptor in humans. Clearly this likelihood will be increased when selective agents are available that can adequately discriminate between the receptor subtypes.
As a first attempt to look at selective ligands, we synthesized and tested several newly reported non-imidazole H3receptor antagonists (Ganellin et al., 1998; Schwartz et al., 1998). One such compound, 4-(3-piperidin-1-yl-propoxy)-benzonitrile, had been reported to be a high-affinity H3 antagonist (Schwartz et al., 1998). We found that 4-(3-piperidin-1-yl-propoxy)-benzonitrile was a high-affinity H3 antagonist (K I = 25 nM) but inactive at GPCR105. To date, we have found a similar result for all non-imidazole H3 antagonists synthesized and tested (not shown). This is not surprising because one would expect that compounds derived from histamine (imidazole-based) would have a higher probability of being relatively equally recognized by highly related histamine-binding receptors such as H3and GPCR105 and thus be nonselective. Non-imidazole H3 antagonists are no longer related to the endogenous common substrate, histamine, and thus would most likely have a higher probability of being selective. This presumption, however, awaits proof through careful chemical development and future structure-activity relationship analysis. Assuming it proves to be correct, one can predict that there is a good probability of identifying selective, non-imidazole GPCR105 ligands.
The tissue expression profile of GPCR105 is rather unique. Our initial examination of expression in cDNA libraries from different tissue sources suggested abundant expression in bone marrow. This was confirmed by RNase protection assay where bone marrow appeared to be the highest site of expression. However, expression was seen in a variety of tissues including spleen, small intestine, and stomach. Bone marrow stem cells give rise to numerous lymphoid and myeloid cell types, including eosinophils. Since Raible et al. (1994) identified a putative novel histamine receptor on eosinophils whose pharmacological profile resembles GPCR105, we examined expression of GPCR105 on human eosinophils and an eosinophil-like subline of HL-60 promyelocytic cells. The HL-60 clone 15 subline has been shown to take on eosinophil characteristics when stimulated with butyric acid and interleukin-5 (Fischkoff, 1988; Tiffany et al., 1995). Indeed, human eosinophils did show significant expression of GPCR105. Similarly, HL-60 clone 15 cells had slight expression of GPCR105, but when differentiated with butyrate/IL-5, showed a robust expression of GPCR105 that could be detected by Northern blot. This shows that GPCR105 is expressed on human eosinophils. We are currently elucidating the role of GPCR105 on human eosinophils.
As our article was in preparation, an on-line version of an article was published that described the same histamine receptors, but with three changes that resulted in different amino acids at positions 138, 206, and 253 (Oda et al., 2000). Our findings are mostly consistent with theirs with respect to the pharmacology such that histamine is a more potent agonist than (R)-α-methylhistamine and that the receptor binds clobenpropit, thioperamide, and clozapine. As for tissue expression, their work focused on leukocytes, small intestine, and eosinophils. They did not evaluate bone marrow per se. One point of difference is that we detected slight expression in brain whereas they did not detect expression there. Our method of RNase protection is very sensitive and thus may detect low levels of expression. Our PCR-based expression pattern was more consistent with theirs in that brain expression was negligible. We are attempting more rigorous evaluation of brain expression via in situ hybridization. Perhaps the discrepancies could be related to variation within tissue specimens since the receptor is expressed on blood borne cells and may be highly regulated. Another interesting point is that Oda et al. (2000)cotransfected their H4 receptor with Gα15 subunit, which appeared to increase their functional coupling. Despite what appears to be better coupling in their coexpressed system, our findings of clobenpropit as a partial agonist and clozapine as a full agonist are almost identical to theirs. The only difference was a switch in the relative potency of clozapine versus (R)-α-methylhistamine. Perhaps, our lack of efficient coupling could be explained by the lack of an appropriate G-protein in the SK-N-MC cells.
In summary, we have cloned and pharmacologically characterized a novel G-protein-coupled receptor (GPCR105) that is consistent with a novel histamine receptor subtype. Its expression in bone marrow and likely expression on eosinophils promises the elucidation of a potential new role for histamine in allergy and asthma. We have tentatively named this new receptor H4 for the fourth histamine receptor subtype. Although its gene sequence and imidazole-based pharmacology are related to the H3 receptor, the functional role of this receptor is likely to be completely distinct to that of the H3 and thus we felt it should be categorized on its own. We are currently knocking out the gene in mice to identify additional physiological roles for the H4 receptor.
Acknowledgments
We thank Drs. Nigel Shankley, Lars Karlsson, and Wai-Ping Leung for providing insightful discussion.
Footnotes
- Received October 12, 2000.
- Accepted December 27, 2000.
-
Send reprint requests to: Timothy W. Lovenberg, The R. W. Johnson Pharmaceutical Research Institute, 3210 Merryfield Row, San Diego, CA. E-mail: tlovenbe{at}prius.jnj.com
Abbreviations
- ATCC
- American Type Culture Collection
- GPCR
- G-protein-coupled receptor
- PCR
- polymerase chain reaction
- UTR
- untranslated region
- RACE
- rapid amplification of cDNA ends
- RPA
- RNase protection assay
- bp
- base pair(s)
- The American Society for Pharmacology and Experimental Therapeutics