Abstract
Cardiac function is regulated by many hormones and neurotransmitters that exert their physiological effects through the activation of G protein-coupled receptors (GPCRs). Identification of new GPCRs that might display a specific pattern of expression within the heart and differentially regulate specific cardiac functions represents an important issue for the development of new drugs. Indeed, highly targeted receptors represent only a small percentage of known GPCRs. Here, we quantified the expression of 395 endoGPCRs (all GPCRs excluding taste and odorant receptors) in male mouse right and left atria and ventricles by using high-throughput real-time reverse-transcriptase polymerase chain reaction (RT-PCR) and focused on the 135 most highly expressed transcripts. No cardiac functional data are available for almost half of these receptors; therefore, linking GPCR expression patterns to cardiac function has allowed us to provide new insights into the possible function of some of these receptors. Indeed, ventricles and atria are both contractile; however, the latter, and especially the right atrium, are central to the generation and regulation of cardiac rhythm. Accordingly, the right atrium exhibited the most specific signature, whereas the majority of GPCRs found in ventricles were evenly expressed in both the right and left chambers. RT-PCR data were confirmed at the protein level for six selected transcripts. Furthermore, we provide new data showing that, as suggested by our repertoire, the metabotropic glutamate receptor 1b is expressed and is functional in ventricular cardiac myocytes. This is the first report describing GPCRs in the four cardiac chambers and may assist in the identification of therapeutic targets.
The four cardiac chambers display morphological and functional differences. They can be distinguished on the basis of contractile properties, rhythm generation, and response to neurohumoral stimulation (Brown et al., 1991; Barth et al., 2005; Narolska et al., 2005). Contractile activity of the heart ensures that blood flow is adapted to the needs of the organism. Left ventricular performance depends on the chamber's capacity to fill up (diastolic property) and to empty (systolic property), and these properties are altered in pathological conditions, the final consequence being heart failure. To trigger the contractile activity, cardiac automaticity is generated by a set of specialized pacemaker cells forming the sinoatrial node (SAN) (Mangoni and Nargeot, 2008). Compared with myocytes of the working myocardium, pacemaker SAN cells are devoted to the generation of an electrical oscillation rather than having a contractile activity. At the molecular level, heart chamber heterogeneity is reflected by the expression patterns of different sets of genes such as transcription factors (Tabibiazar et al., 2003) and ion channels (Marionneau et al., 2005). The heart expresses GPCRs (Regard et al., 2008), which also play key roles in cardiac physiology and performance (Tang and Insel, 2004). One would expect that these receptors display a heterogeneous or chamber-specific distribution. How these receptors are expressed in specific regions of the heart and how they function in physiological conditions are important issues to be addressed.
GPCRs constitute the largest family of transmembrane signaling molecules (Lefkowitz, 2004). They can be activated by a variety of extracellular signals, including neuropeptides, chemokines, biogenic amines, hormones, lipid-derived mediators, proteases, light, taste, and smell. Upon ligand binding, GPCRs transduce these signals into intracellular responses that regulate cell function via the heteromeric G proteins. A large fraction of GPCRs are orphan receptors, for which the corresponding ligands have not yet been identified (Lefkowitz, 2004; Tang and Insel, 2004; Chung et al., 2008). Given the major roles of GPCRs in cardiac regulation, one can wonder how many different GPCRs exist in the heart and what is their physiological significance. Many GPCRs are key targets for maintaining cardiovascular homeostasis because they initiate signaling cascades that regulate intracellular Ca2+ (Ca2+i), a key messenger in processes such as excitation-contraction coupling and gene transcription (Tang and Insel, 2004). It is noteworthy that the adrenergic receptors (ARs) have been extensively studied and are the targets for many drugs used in cardiac diseases. β1-AR, the predominant AR subtype in heart, is coupled to Gs and mediates the positive inotropic effect of sympathetic stimulation by increasing myocyte Ca2+i via L-type calcium channels. On the other hand, GPCRs preferentially coupled to Gq, such as angiotensin II (AT1) and endothelin (ETA) receptors, increase Ca2+i through a phospholipase C-dependent inositol triphosphate production and are major targets for antagonists used in cardioprotection. Identification of new GPCRs that might display a specific pattern of expression within the heart and differentially regulate specific cardiac functions also represents an important issue for the development of new drugs. Indeed, approximately 30 to 40% of prescriptions drugs act on GPCRs, but these highly targeted receptors represent only a small percentage of known GPCRs (Hopkins and Groom, 2002).
In the present study, we used quantitative, real-time RT-PCR to establish the complete endoGPCR repertoires (all GPCRs excluding taste and odorant receptors) of the four mouse cardiac chambers. This study gives a global and regional view of the expression of new GPCRs in the heart and represents a valuable resource to improve our understanding of cardiac physiology and to identify new pharmacological targets.
Materials and Methods
Detailed descriptions of the materials and methods used are available in the Supplementary materials. All animal procedures were performed according to the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996).
RNA Extraction and Reverse Transcription. Twenty hearts were dissected from 8-week-old C57BL/6 male mice (Charles River, Margate, Kent, UK). Total RNAs were prepared from pooled cardiac chambers using the TRIzol method according to the manufacturer's instructions (Invitrogen, Carlsbad, CA): right atria (RA), including the SAN; left atria (LA); and right (RV) and left ventricles (LV). RNAs were treated with DNase I RNase-Free (Ambion, Austin, TX) and purified by LiCl precipitation. Three independent reverse transcriptions (RTs) were performed using 3.5 μg of total RNA with random primers and the Moloney murine leukemia virus reverse transcriptase (Invitrogen). Identical reactions were also set up in the absence of reverse transcriptase to eliminate the possibility of genomic contamination.
Real-Time RT-PCR. The expression levels of genes were determined by quantitative PCR using SYBR Green and an ABI Prism 7500 machine (Applied Biosystems, Foster City, CA). The 402 genes selected for quantification encode 395 endoGPCRs and 7 associated proteins: Ramps (Ramp1, Ramp2, and Ramp3) and Crcp that are known to alter the pharmacology of B family GPCRs, as well as secreted inhibitors of the frizzled family (Frza/Sfrp1, Sfrp2, and Frzb/Sfrp3). The multistep process used to select GPCR sequences is described in the Supplementary Methods. The entire set of murine endoGPCR primers is available in the GEO database (see Supplementary Materials). Primer pairs did not span introns and were validated using mouse genomic DNA as a template. Primers giving at least 90% efficacy as well as single amplicons with an appropriate melting temperature and size were selected and further validated on a pool of cDNAs from different mouse tissues. No amplicons were generated from RT-minus samples. Experiments were performed on 1/700th of the cDNA product using 1× SYBR Green PCR Master Mix TaqMan PCR buffer from Applied Biosystems and 300 nM concentration of each primer pair. The cycling conditions included a hot start for 10 min at 95°C, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min and a final cycle of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Data were analyzed using the threshold cycle (Ct) relative quantification method. Three housekeeping genes (HKGs) among the eight tested, hypoxanthine guanine phosphoribosyl transferase (HPRT), β2 microglobuline (β2M), and β-glucuronidase (GUS), were chosen to normalize the results using the geNorm method (Vandesompele et al., 2002). The expression level of each gene was normalized according to the formula 2-[Ct (gene)-arithmetic mean (Ct(HPRT),Ct(β2M), Ct(GUS))] × 100. Results are expressed as the percentage relative to the geometric average of the expression levels of the three selected HKGs.
Two-Way Hierarchical Data Analysis. Classification was performed with a two-way hierarchical clustering using the Cluster and Mapletree software packages. The input consisted of the average gene expression levels measured in the four cardiac chambers coming from three independent RT-PCRs measured in monoplicate (Supplementary Table 1). Genes were filtered according to two criteria: 1) the level of gene expression, all genes whose expression was less than 1% of the geometric mean of the expression levels of the three HKGs in the four chambers were excluded; and 2) differential expression (Eisen et al., 1998), evidenced by one-way ANOVA (p < 0.05) followed by the Tukey post hoc test for contrasts. ANOVA p values were corrected for multiple testing using the Benjamini and Hochberg method. Of the 135 genes loaded, 89 genes passed. Expression levels of these genes were centered against the mean and log-transformed. Average linkage clustering was applied with uncentered correlation using the Cluster program (Eisen et al., 1998). Clustering was visualized using the Mapletree program.
Western Blot Analysis. Membrane proteins were extracted from RA, LA, RV, LV, and control tissue samples from 20 8-week-old C57BL/6 male mice (Charles River). Samples were fractionated on 10% polyacrylamide gels in the presence of SDS and transferred onto nitrocellulose membranes for immunodetection.
Isolation of Adult Mouse Cardiomyocytes. Ventricular myocytes from wild-type and mGluR1(-/-) mice (B6.129P2-Grm1tm1Dgen/J strain, provided by The Jackson Laboratory, Bar Harbor, ME) were isolated by perfusing whole hearts on a Langendorff apparatus with a solution containing collagenase type II. Left ventricular myocytes were plated on laminin-coated glass-bottomed 35-mm dishes (Willco Wells, Amsterdam, the Netherlands) and allowed to adhere for 2 h.
Calcium Imaging. Ventricular myocytes were loaded with fura-2-acetoxymethyl ester (Invitrogen). Imaging of intracellular calcium changes was performed using the MetaFluor Imaging System (Molecular Devices, Sunnyvale, CA).
Statistical Analysis. Gene expression patterns were analyzed as described above. For calcium-imaging experiments, ANOVA followed by the Tukey post hoc test was used to compare cell responses to agonists and antagonists. Results are expressed as mean ± S.D. or S.E.M. as indicated in the figure legends. A p value < 0.05 was considered to be statistically significant.
Results
Expression of GPCRs in the Heart. To define the full endoGPCR repertoires of the four cardiac chambers, high-throughput real-time RT-PCR was performed on RNAs obtained from the pooled right atria, left atria, right ventricles, and left ventricles of 20 C57BL/6 adult male mice. This method offers improved sensitivity, specificity, and reproducibility over conventional genome-wide expression profiling tools such as DNA microarrays. The whole data set, comprising 395 GPCRs and 7 GPCR-associated proteins, is presented in Supplementary Table 1. To facilitate the analysis, we focused on the 135 most abundantly expressed transcripts, including 128 GPCRs and 7 GPCR-associated proteins, whose expression levels were more than 1% relative to 3 selected HKGs (see Materials and Methods). This arbitrary cutoff level does not imply that transcripts with <1% expression relative to the HKGs are not physiologically relevant. Cross-contamination between chambers was excluded by quantifying the expression of specific markers, as illustrated in the Supplementary Fig. 1.
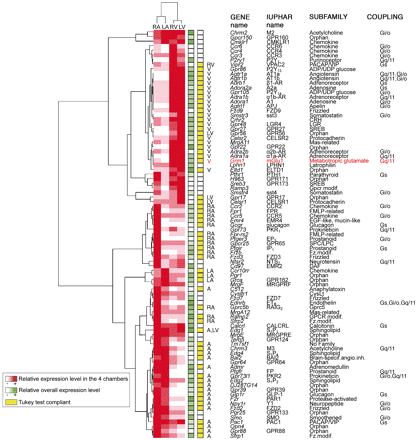
To identify genes with an expression level statistically different in at least one chamber, we used an ANOVA (p < 0.05) on the expression levels of these 135 transcripts. Eighty-four GPCRs and five GPCR-associated proteins were differentially expressed in at least one chamber. To illustrate the quality of the RT-PCR data, we have represented results obtained for the entire Frizzled subfamily (Supplementary Fig. 2). Indeed, data for this subfamily encompass the full range of expression levels and illustrate receptors more specifically expressed in the right atrium (Fzd3), in atria (Fzd2), and in ventricles (Fzd9). To identify transcripts expressed more specifically in supraventricular versus ventricular cardiac tissue, data were hierarchically clustered (Fig. 1, see Materials and Methods). This method resulted in a dendogram that classifies transcripts based on their differential regional expression pattern. We then used the supplementary post hoc Tukey test (p < 0.05) to perform pair-wise comparisons of means to evaluate differences in atrial versus ventricular myocardium and in left versus right heart (Fig. 1). The 44 receptors and 2 associated proteins that were not differentially expressed were considered as uniformly expressed throughout the heart (Fig. 2).
GPCRs Preferentially Expressed in Atria. In both atria, we found a high expression of the M3 subtype of acetylcholine (muscarinic) receptor, whereas the M2 receptor (representing more than 90% of total muscarinic mRNA) was highly expressed throughout the heart but with a significantly lower level in the right atrium. This is consistent with previous studies showing that acetylcholine may exert a negative chronotropic effect via the M3 receptor (Wang et al., 2007). In addition, other receptors highly expressed in atria included the adrenomedullin Admr, sphingolipid S1P2 and S1P4, prokineticin PKR2, protease-activated PAR1, neuropeptide Y1, pituitary adenylate cyclase-activating peptide PAC1, and Frizzled FZD2 receptors. We also identified the Frizzled receptor regulatory protein secreted frizzled related protein 1 (Sfrp1) and the glucagon-like peptide receptor GLP-1 in both atria. It is noteworthy that the GLP-1 receptor has not been characterized in the heart, but brain GLP-1 has been shown to control the heart rate (Cabou et al., 2008).
The right atrium revealed high levels of transcripts for many receptors, including the glucagon, N-formylpeptide-related FPR and Fpr-rs2, chemokine CCR2 and CCR5, sphingosylphosphorylcholine/lysophosphatidylcholine GPR65, prostanoid IP1, and Frizzled FZD3 receptors. The Admr receptor-associated protein, Ramp2, and Frizzled receptor-associated proteins Sfrp2 and Frzb were also highly expressed in this chamber. The right atrium also highly expressed receptors with no known cardiac function, including the epidermal growth factor-like, mucin-like EMR4 receptor. Fewer GPCRs were specific to the left atrium: the Chemokine Ccr10rr receptor and two orphan receptors, Pgr1 and GPR162.
GPCRs Preferentially Expressed in Ventricles. Thirteen classic cardiac receptors were expressed evenly in both ventricles, including angiotensin, adenosine, membrane-bound P2, and adrenergic receptors. As reported previously (Woodcock et al., 2008), adrenergic receptors were the most represented with the following relative abundance: α1b>α1a>α2b and β1>β2, with β2 being uniformly expressed throughout the heart. We also found transcripts for the apelin APJ and the corticotropin-releasing hormone Crhr2 receptors. In addition, we evidenced a lot of receptors for which, to our knowledge, no role in heart has so far been described. They included receptors for the protocadherin, super-conserved receptor expressed in brain, and Mas-related receptor subfamilies. Finally, we identified the metabotropic glutamate mGlu1 receptor transcript (called mGluR1 throughout the text) that we further analyzed in cardiomyocytes (see below).
The expression patterns varied far less between ventricles than between atria. Indeed, the right ventricle was characterized only by a higher expression of the vasoactive intestinal peptide/pituitary adenylate cyclase-activating peptide VPAC2 receptor. The left ventricle strongly expressed two orphan receptors, GPR17 and GPR56, and to a lesser extent the protocadherin CELSR1 receptor.
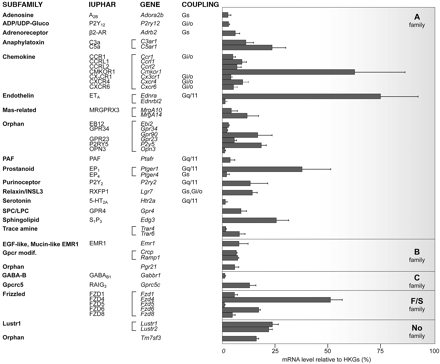
GPCR Transcripts Uniformly Expressed in Mouse Heart. GPCRs equally expressed in the four cardiac chambers of mouse heart are shown in Fig. 2. Several genes displayed a remarkably high expression level in all four heart chambers, suggesting that they may play a prominent role in heart physiology. Indeed, the endothelin ETA receptor was the most abundantly expressed overall and is well known for modulating cardiac contractile function (Piuhola et al., 2003). Quite surprisingly, little information is available concerning the physiological cardiac function of many of the most abundantly expressed receptors: the anaphylatoxin C5a, prostanoid EP1, sphingolipid S1P3, frizzled FZD4, chemokine CMKOR1, and lung seven-transmembrane receptors Lustr1 and Lustr2.
Among the less abundant receptors, we found the β2-AR, the purinoceptor P2Y2, the most highly represented pyrimidine receptor in human cardiomyocytes (Wihlborg et al., 2006), and receptors from the anaphylatoxin, frizzled, chemokine, and relaxin subfamilies. Among GPCR-associated proteins, CGRP-receptor component protein (Crcp) and receptor-activity modifying protein 1 (Ramp1) were highly expressed throughout the heart. A substantial number of receptors whose presence and/or function have not been reported in the heart were also uniformly expressed: the Mas-related MrgA14, sphingosylphosphorylcholine/lysophosphatidylcholine GPR4, trace amine Trar6 (also called Taar6), epidermal growth factor-like/mucin-like EMR1, Gprc5 RAIG3, and orphan (Gpr90, P2RY5, and Tm7sf3) receptors. It is noteworthy that other receptors with well characterized functions in the heart, including the adenosine A2B, serotonin 5-hydroxytryptamine2A, and GABAB1 receptors were uniformly expressed, but at a lower level.
Expression profiles of endoGPCRs differentially expressed in at least one of the four cardiac chambers. A two-way hierarchical clustering was applied to the 89 differentially expressed genes. The relative expression level is red color-coded for each gene in the four cardiac chambers. The entire dendogram is shown on the left. Boxes filled with shades of green indicate the highest expression level among the four cardiac chambers of the considered gene compared with that of the others. In addition, if the cluster is compliant with the Tukey post hoc test, genes are marked with a yellow box, next to which the chamber(s) with preferential expression is indicated (significantly high values are underlined in Supplementary Table 1). Gene names, International Union of Basic and Clinical Pharmacology names, preferential G-protein coupling (available at http://www.iuphar-db.org/index.jsp), and subfamilies are mentioned on the right. RV, right ventricle; A, atria; V, ventricles.
Expression levels of GPCRs uniformly expressed throughout the heart. RT-PCR results from the left ventricle (pool of 20 mice) are presented for the 46 uniformly expressed genes. Results show gene expression levels ± S.D. as a percentage of the geometric average of the expression level of three HKGs (see Supplementary Data). Subfamilies, International Union of Basic and Clinical Pharmacology nomenclature, preferential G-protein coupling, and gene names are indicated.
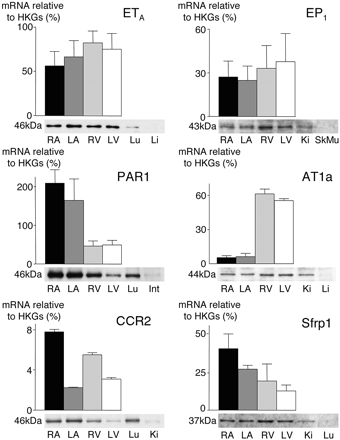
Western Blotting Confirms GPCR Expression. Western blot experiments were conducted to relate protein expression to that of mRNA for selected genes. Equal amounts (27 μg) of membrane extract protein obtained from 20 mice were loaded on SDS/PAGE gels. In each case, we used a positive and negative control tissue. Figure 3 shows original blots for RA, LA, RV, and LV below the corresponding QRT-PCR data. These experiments confirmed the expression of the specific receptor proteins in the heart. The protein levels were consistent with the corresponding transcriptional data for the ETA, EP1, PAR1, and AT1a receptors; the CCR2 receptor and Sfrp1 protein distributions correlated to a lesser extent with QRT-PCR data.
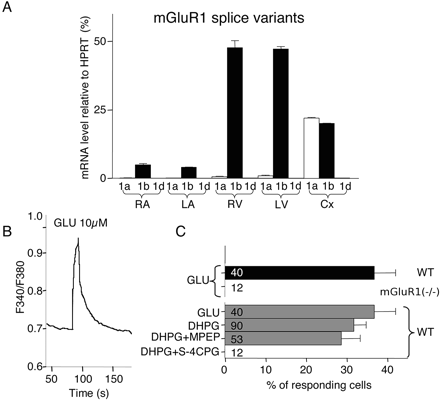
Functional Analysis of the mGluR1b Glutamate Receptor. mGluR1 was the only metabotropic glutamate receptor transcript in our mouse repertoire as opposed to the rat heart (Iglesias et al., 2007). We were unable to detect any specific band corresponding to the mGluR1 protein in the mouse heart (data not shown). Because alternative splicing generates different variants of mGluR1, we designed specific QRT-PCR primers to quantify these variants in the four cardiac chambers, using mouse brain cortex as a control (Fig. 4A). We confirmed that mGluR1b is the only variant highly expressed in mouse heart (Zhu et al., 1999); this result can explain our negative Western blot data, because the available anti-mGluR1 antibodies recognize only the mGluR1a variant.
We then checked the functional expression of the mGluR1b receptor by measuring Ca2+i mobilization in ventricular cardiomyocytes by fluorometric experiments using fura-2. Application of glutamate (GLU), added to the recording solution (10 μM for 1 min), induced a single-peak, nonoscillatory [Ca2+]i increase (Fig. 4B), a hallmark of mGluR1 receptors (Kawabata et al., 1996). A percentage (36 ± 5%) of all cardiomyocytes tested responded to GLU (40 cells from 7 different mice; Fig. 4C), a similar percentage of responding cells was observed for ANGII and endothelin-1 applications (data not shown). No response to GLU was detected in any ventricular myocytes from mGluR1(-/-) mice (12 cells from 5 mice), whereas these same cells were able to respond to ANGII similarly to wild-type cells (data not shown). The response to GLU obtained in wild-type mice resulted from the activation of group I mGluRs because a similar percentage of cells responded to the specific group I agonist (S)-3,5-dihydroxyphenylglycine (DHPG; 32 ± 3%, 90 cells from 14 mice). We next examined the effects of subtype-specific mGluR1 and mGluR5 antagonists (100-fold excess) on DHPG (3 μM)-induced Ca2+i responses (Fig. 4C). Cells were first challenged by DHPG in association with the low-affinity mGluR1 and mGluR5 antagonist S-4-carboxyphenylglycine (S-4CPG) (Brabet et al., 1995) and then, after a wash period of 5 min, by DHPG in the presence of 2-methyl-6-(phenylethynyl) pyridine hydrochloride (MPEP), a specific mGluR5 subtype antagonist (Gasparini et al., 1999). S-4CPG completely inhibited DHPG-induced [Ca2+]i elevations in these myocytes, whereas MPEP had no effect (12 cells from 7 mice). Accordingly, cells stimulated with DHPG in the presence of MPEP responded similarly to cells stimulated with DHPG alone or with glutamate (29 ± 5%, 53 cells from 5 mice). Taken together, these results demonstrate that glutamate and DHPG induced an increase of Ca2+i via the mGluR1b receptor in ventricular cardiomyocytes.
mRNA and protein expression of uniformly and differentially expressed GPCRs in the four cardiac chambers. Western blot analysis in RA, LA, RV, and LV are shown below the corresponding QRT-PCR data. Positive (left) and negative (right) control tissues are shown next to the four chambers. Protein (27 μg) from membrane extracts was loaded into each well. Lu, lung; Li, liver; Ki, kidney; SkMu, skeletal muscle; Int, intestine.
Quantitative and functional analysis of mGluR1 splice variants. A, quantification of mGluR1a, -b, and -d splice variant transcripts in mouse RA, LA, RV, LV, and brain cortex (Cx). mRNA contents were normalized according to HPRT, and the results are expressed as 2-[Ct(mGluR1) - Ct(HPRT)] × 100. Data are expressed as means ± S.E.M. (n = 3). B, representative recording of the variation of [Ca2+]i in response to an application of GLU. C, histograms represent the average number of ventricular myocytes (percentage) per mouse that responded to GLU (10 μM), DHPG (3 μM), DHPG (3 μM) + MPEP (300 μM), and DHPG (3 μM) + S-4CPG (300 μM). Digits indicate the total number of cells studied. Data are expressed as means ± S.E.M. (n = 5-14 mice).
Discussion
We have performed the first genome-wide, comprehensive expression profiling of endoGPCRs within the four cardiac chambers of the adult mouse. Highly expressed transcripts have been classified into two groups, those differentially and those uniformly expressed throughout the heart. To our knowledge, there is no cardiac functional data for almost half of these receptors, including entire subfamilies such as the super-conserved receptor expressed in brain and Mas-related GPCRs.
GPCR Transcripts Differentially Expressed in Mouse Heart. Indeed, ventricles and atria are both contractile; however, the latter, and especially the right atrium, are central to the generation and regulation of cardiac rhythm. These specific roles are consistent with the clustering of receptors from our repertoires into two major groups: those that are preferentially expressed in atria (55%), including a cluster specific to the right atrium, and those preferentially expressed in ventricles (45%; Fig. 1). This partition reflects the fact that atria are central to the neurohumoral regulation of cardiac function and therefore express higher levels of genes involved in receptor signaling than ventricles (Barth et al., 2005). Within these groups, further differential expression patterns were found between atria and less between ventricles. Of atrium-specific GPCRs, 44% were differentially expressed between both atria versus 16% of ventricle-specific GPCRs differentially expressed between both ventricles. No gene seemed to be significantly specific to the left or right side of the heart.
The right atrium exhibited the most specific signature with a lot of new receptors. Indeed, we found that 35% of the atrial-specific receptors were preferentially expressed in the right atrium (versus 9% in the left atrium), suggesting that many of them may be involved in the regulation of cardiac automaticity, because the right atrium contains the sinoatrial node, where the heart beat is initiated. New receptors included the IP1 and EP3 prostanoid receptors. Interestingly, the activation of FP prostanoid receptors, which we found to be highly expressed in both atria, has been shown to increase the heart rate (Takayama et al., 2005). Admr, receptor of the potent vasodilator adrenomedullin, and its chaperone protein Ramp2 were both highly expressed in the right atrium, whereas the related CALCRL receptor, together with its chaperone protein Ramp1, were expressed throughout the heart. This result is consistent with data showing that adrenomedullin exerts Ca2+-dependent-positive inotropic effects in human atria and, to a lesser extent, in ventricular myocardium (Bisping et al., 2007). Only the chemokine Ccr10rr receptor and two orphan receptors, Pgr1 and GPR162, constituted the signature of the left atrium.
The vast majority of GPCRs found in ventricles were evenly expressed in both the right and left ventricles. Many of them are involved in the regulation of heart contractility, which is under the control of the sympathetic nervous system via the release of norepinephrine and epinephrine that activate cardiomyocyte ARs. We found that β1-AR and α1b-AR were the predominant subtypes in both mouse ventricles as observed previously in rodents (Woodcock et al., 2008). The role of the β1-AR subtype has been widely studied in terms of the regulation of both inotropic and chronotropic contractile function and in cardiac hypertrophy. Recent studies now provide support for an important role of α1-AR in maintaining cardiac contractility under pathological conditions, such as ischemia or pathological hypertrophy, that are associated with decreased β1-AR function (Woodcock et al., 2008). Among novel receptors, we show a high and specific expression in both ventricles of the apelin receptor (APJ), structurally related to the AT1 receptor. This receptor, whose endogenous ligand is the vasoactive peptide apelin, does not bind ANGII. A recent review highlights the diverse roles played by the apelin/APJ receptor in cardiovascular function including its contractile action (Chandrasekaran et al., 2008).
GPCR Transcripts Uniformly Expressed in Mouse Heart. Among the uniformly expressed receptors, we found well characterized receptors such as the adenosine, adrenergic, and endothelin receptors. We also evidence some new receptors with emerging functions such as receptors for trace amines (Frascarelli et al., 2008) or chemokines (Husberg et al., 2008), the latter best known as inflammatory mediators. High expression levels of these receptors in normal heart raises the question of their role under physiological conditions. Recent studies have shown that the CXCL12 receptor CXCR4 found in adult cardiac myocytes is involved in postinfarction left ventricular remodelling and modulates contractility (Pyo et al., 2006). It is noteworthy that we found CXCR4 to be highly expressed throughout the heart, along with CMKOR1 (also called Cxcr7), in agreement with a study showing that the two heterodimerize (Sierro et al., 2007). Another group of receptors ubiquitously expressed in the adult heart was that known to have an important physiological role during embryonic development (i.e., frizzled receptors) and their associated proteins Sfrps and Frzb (van de Schans et al., 2008). The presence of these receptors in adult mouse implies that their role may not be restricted to heart development. Indeed, the overexpression of Sfrp1 is able to reduce infarct size and improve cardiac function (Barandon et al., 2003). We can hypothesize that some of these GPCRs might be present to adapt to the modifications occurring during cardiac remodelling.
Transcriptome Validation. For any receptor, knowledge about the mRNA/protein expression ratio is important. A high-throughput technique equivalent to QRT-PCR is not yet widely available for quantifying proteins. In addition, we were limited by the number of available and validated antibodies against GPCRs. Hence, we selected a group of five receptors (and one GPCR-associated protein) that we believed were representative of our transcriptome. Indeed, they differ in respect to their physiological role in the heart, transcript distribution (uniform versus differential), or their overall level of expression. For these six genes, we confirmed the expression of the corresponding proteins, particularly for the CCR2 receptor and Sfrp, both poorly characterized in healthy heart. In addition, our results revealed that transcript levels in the four chambers were largely reflected at the protein level, especially for the most highly expressed transcripts. mRNA and protein levels do not systematically correlate as a result of tissue and/or cell type-specific post-transcriptional regulations (Gygi et al., 1999).
Given that heart is a heterogeneous tissue made up of not only myocytes but also endothelial cells, fibroblasts, and intracardiac neurons, it is essential to assess protein function at the cellular level. Our demonstration that the metabotropic glutamate receptor 1b, evidenced at the messenger level in whole tissue, was present and functional in ventricular cardiomyocytes, validates our repertoire and is also of intrinsic interest. The main known physiological function of glutamate in the heart, where it has a high intra- (millimolar range) to extracellular (<1 μM range) concentration ratio, is to regulate energy metabolism by maintaining the levels of Krebs cycle intermediates, an effect independent of glutamate receptor activation. This process, known as anaplerosis, has been proposed to be the basis for the cardioprotective effects of glutamate (Langenberg et al., 2001). On the other hand, glutamate may also have toxic effects at supraphysiological concentrations (Gill et al., 2007). Indeed, arrhythmias and other cardiovascular symptoms have been reported in humans intoxicated with monosodium glutamate from food. The role of glutamate via its own receptor remains unknown, and there are some discrepancies regarding the isoform and even the presence of mGluR1 in the heart. An immunohistochemistry study has reported the presence of mGluR5 in intercalated disks of myocytes, especially in the ventricles, and of mGluR2/3 and mGluR1 in various cell types of rat atrium (Gill et al., 1999). A similar study showed positive staining for mGluR1 and mGluR5 in both atrial and ventricular human cardiomyocytes (Gill et al., 2007). Another study has reported the presence of mGluR1 and mGluR5 in whole rat heart homogenates, as well as demonstrating a functional coupling of these receptors to phospholipase C (Iglesias et al., 2007). Although a recent study did not detect any metabotropic glutamate receptor in mouse heart (Regard et al., 2008), we and Zhu et al. (1999) found a high expression of mGluR1b in mouse heart. In addition, we provide new evidence for a physiologically relevant role for mGluR1 by showing that functional receptors are present in isolated ventricular myocytes and are able to induce an increase in intracellular calcium, probably by an inositol triphosphate-mediated response via Gαq. Signal transduction through mGluR1b in cardiomyocytes could have physiological and pathological relevance and needs to be investigated further. Indeed, glutamate is the main amino acid known to be released during cardiac ischemia (Kennergren et al., 1999).
In conclusion, regional GPCR gene expression patterns should help to provide new insights into the functional differences between chambers and roles of individual and groups of both well known and uncharacterized receptors in the heart. We also show the distribution of GPCR-associated proteins such as RAMPs, Crcp, and secreted inhibitors of the Frizzled family that form complexes with receptors. Studying the distribution of key GPCR-associated proteins could provide information on likely receptor coupling and functional differences between chambers. Overall, this global approach should contribute to unraveling the complex cardiac GPCR signaling networks and identifying new pharmacological targets.
Acknowledgments
We are grateful to J. Bockaert, J.P. Pin and his group, and the staff of the Department of Physiology (Institut de Génomique Fonctionnelle) for constructive and supportive discussions. We also thank C. Sarrauste de Menthière, A. Turner-Madeuf, M. Gien-Asari, P. Atger, A. Cohen-Solal, D. Addou, and E. Gavois for their excellent technical assistance.
Footnotes
-
The work was supported by the Association Française contre les Myopathies [Grant 12335] and La Fondation de France [Grant 032144].
-
ABBREVIATIONS: SAN, sinoatrial node; ANGII and AT1, angiotensin II; β2M, β2 microglobuline; DHPG, (S)-3,5-dihydroxyphenylglycine; GLU, glutamate; GPCR, G protein-coupled receptor; GUS, β-glucuronidase; HKG, housekeeping gene; HPRT, hypoxanthine guanine phosphoribosyl transferase; LA, left atrium; LV, left ventricle; MPEP, 2-methyl-6-phenylethynyl; PCR, polymerase chain reaction; RA, right atrium; RT, reverse transcription; RT-PCR, reverse transcription-polymerase chain reaction; RV, right ventricle; S-4CPG, S-4-carboxyphenylglycine; AR, adrenergic receptor; ETA, endothelin; ANOVA, analysis of variance; QRT-PCR, quantitative reverse transcription-polymerase chain reaction.
-
↵
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material. - Received December 12, 2008.
- Accepted February 19, 2009.
- The American Society for Pharmacology and Experimental Therapeutics