Abstract
Specialized proresolving mediators (SPMs) and their cognate G protein-coupled receptors are implicated in autoimmune disorders, including chronic inflammation, rheumatoid arthritis, systemic scleroderma, and lupus erythematosus. To date, six G protein-coupled receptors (GPCRs) have been paired with numerous endogenous and synthetic ligands. However, the function and downstream signaling of these receptors remains unclear. To address this knowledge gap, we systematically expressed each receptor in a human embryonic kindney 293 (HEK293)-Flp-In-CD8a-FLAG cell system. Each receptor was pharmacologically characterized with both synthetic and putative endogenous ligands across different signaling assays, covering both G protein-dependent (Gs, Gi, and Gq) and independent mechanisms (β-arrestin2 recruitment). Three orphan GPCRs previously identified as SPM receptors (GPR 18, GPR32 and GPR37) failed to express in HEK 293 cells. Although we were unsuccessful in identifying an endogenous ligand for formyl peptide receptor 2 (FPR2)/lipoxin A4 receptor (ALX), with only a modest response to N-formylmethionine-leucyl-phenylalanine (fMLP), we did reveal clear signaling bias away from extracelluar signal-related kinase (ERK) 1/2 phosphorylation for the clinically tested agonist N-(2-{[4-(1,1-difluoroethyl)-1,3-oxazol-2-yl]methyl}-2H-1,2,3-triazol-4-yl)-2-methyl-5-(3-methylphenyl)-1,3-oxazole-4-carboxamide (ACT-389949), adding further evidence for its poor efficacy in two phase I studies. We also identified neuroprotectin D1 as a new leukotriene B4 receptor 1 (BLT1) agonist, implying an alternative target for the neuroprotective effects of the ligand. We confirmed activity for resolvin E1 (RvE1) at BLT1 but failed to observe any response at the chemerin1 receptor. This study provides some much-needed clarity around published receptor-ligand pairings but indicates that the expression and function of these SPM GPCRs remains very much context-dependent. In addition, the identification of signaling bias at FPR2/ALX may assist in guiding design of new FPR2/ALX agonists for the treatment of autoimmune disorders.
SIGNIFICANCE STATEMENT To our knowledge, this is the first study to comprehensibly show how several natural mediators and synthetic ligands signal through three specialized proresolving mediator GPCRs using multiple ligands from different classes across four-six endpoint signaling assays. This study discovers new ligand pairings, refutes others, reveals poly-pharmacology, and identifies biased agonism in formyl peptide receptor 2/lipoxin A4 receptor pharmacology. This study highlights the potential of these receptors in treating specific autoimmune diseases, including rheumatoid arthritis, systemic scleroderma, and systemic lupus erythematosus.
Introduction
Resolution of inflammation involves specialized proresolving mediators (SPMs) that tightly regulate inflammation and tissue repair. After the proinflammatory phase, where polymorphonuclear leukocytes (PMNs) are recruited to the injury site, resolution describes the process of PMN apoptosis and efferocytosis by infiltrating macrophages. During this phase, a range of SPMs are released from apoptotic PMNs and recruit macrophages that drive resolution (Park et al., 2020). Chronic inflammation results from inadequate or dysregulated resolution (Fullerton and Gilroy, 2016); however, the role that individual SPMs and their target G protein-coupled receptors (GPCRs) play in these processes remains to be fully elucidated.
SPMs are a diverse set of lipid mediators synthesized from polyunsaturated fatty acids and include E- and D-series resolvins, protectins, maresins, and lipoxins (Chiang and Serhan, 2017). SPMs and the proresolving proteins chemerin, annexin A1 (AnxA1), and serum amyloid A (SAA) are reported to mediate their effects via six G protein-coupled receptors (GPCRs): leukotriene B4 receptor 1 (BLT1), chemerin receptor 1 (chemerin1), N-Formyl peptide receptor 2/lipoxin A4 receptor (FPR2/ALX), and the orphan receptors GPR 18, GPR32, and GPR37 (Park et al., 2020).
This system is highly complex, reportedly exhibiting both ligand poly-pharmacology (a ligand interacting with multiple target receptors) and receptor promiscuity (a receptor that binds multiple endogenous ligands) (Arienti et al., 2019; Park et al., 2020). These SPM-GPCR pairings have been determined using a range of different techniques and assay formats.
BLT1 is a well-established target of leukotriene B4 (LTB4) (Yokomizo et al., 1997), which when activated inhibits cAMP accumulation (Yokomizo et al., 1997; Arita et al., 2007), generates calcium influx (Yokomizo et al., 1997; Ito et al., 2002), and drives phosphatidylinositol-3 kinase (PI3K)/Akt activation and nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) inhibition (Arita et al., 2007). It has been suggested that resolvin E1 (RvE1) is a partial agonist with low nanomolar affinity for BLT1, capable of antagonizing LTB4 (Arita et al., 2007). In vivo observations of RvE1 activity at BLT1 and chemerin1, which displays higher affinity for RvE1 than chemerin itself (Arita et al., 2007), produced mixed results. RvE1-mediated increases in phagocytosis by macrophages (Ohira et al., 2010) was abolished in Cmklr1 knockout mice (Laguna-Fernandez et al., 2018), whereas RvE1-mediated inhibition of PMN recruitment in a zymosan-A-induced peritonitis model was significantly reduced in Ltb4r1 knockout mice (Arita et al., 2007).
FPR2/ALX, which was deorphanized by its high sequence homology with FPR1 and activation by the low affinity formyl peptide N-formylmethionine-leucyl-phenylalanine (fMLP) (Ye et al., 1992), is reportedly a target of multiple SPMs, including lipoxin A4 (LXA4), SAA, LL37, AnxA1, and Resolvin D1 (RvD1) (Krishnamoorthy et al., 2010).
The receptor is also amenable to the peptides WKYMVm and WKVMVM and the investigational small molecule, N-(2-{[4-(1,1-difluoroethyl)-1,3-oxazol-2-yl]methyl}-2H-1,2,3-triazol-4-yl)-2-methyl-5-(3-methylphenyl)-1,3-oxazole-4-carboxamide (ACT-389949) (Maciuszek et al., 2021). In the context of this study, β-arrestin2-mediated FPR2/ALX internalization by ACT-389949 is thought to underpin loss of therapeutic efficacy in phase I clinical trials (NCT02099071 and NCT0209920; Stalder et al., 2017), suggesting that clinical development of FPR2/ALX ligands with bias away from β-arrestin2 recruitment will improve therapeutic efficacy.
The breadth of SPM ligand and GPCR target pairings, coupled with the different in vitro and in vivo techniques used indicates a need for clearer, more consistent determination of these designations. In this study, we performed a systematic pharmacological exploratory characterization of 12–15 commercially available SPM ligands from multiple classes (including resolvins, protectins, lipoxins, peptides, lipids, and synthetic molecules) at three SPM GPCRs stably expressed in human embryonic kidney 293 (HEK 293) cells across assays targeted at key signaling pathways: cAMP accumulation (stimulation and inhibition), ERK1/2 phosphorylation, [35S]-guanosine 5′-(γ-thio)triphosphate (GTPγ35S) binding, calcium, and β-arrestin2 recruitment.
We both confirm existing ligand-receptor pairings, including RvE1 as a low potency agonist of BLT1, and present evidence that questions current pairings. For example, we detected no activity of RvE1 at chemerin1. We confirm that fMLP is a weak agonist at FPR2/ALX but were unable to confirm activity of other proposed endogenous ligands at this receptor. More interestingly, we identify new pairings, including neuroprotectin D1 (NPD1)—recently identified as a GPR37 ligand (Bang et al., 2018; Bang et al., 2021)—as an agonist of BLT1. Finally, using bias analysis, we determined that the FPR2/ALX agonist ACT-389949 is strongly biased away from ERK1/2 phosphorylation relative to the synthetic peptide WKYMVm, providing further context for its limited clinical efficacy.
Materials and Methods
Cell Line Generation and Culture
Stably expressing cell lines for each receptor (FPR2/ALX, BLT1, chemerin1, GPR18, GPR32, and GPR37) were generated by transfecting HEK293 cells with cDNA encoding for HEK293-Flp-In-CD8a-FLAG-‘GPCR’, at a concentration of 2.4 μg and a cDNA:transfection reagent ratio of 1:2.5, using lipofectamine (LTX; ThermoFisher Scientific, Waltham, MA). Clonal populations were identified and maintained in normal growth medium (Dulbecco’s modified eagle’s medium; DMEM; Thermo Fisher Scientific, 10% FBS (Thermo Fisher Scientific, Waltham, MA) at 37°C with 5% carbon dioxide (CO2). Cells were periodically selected using 200 μg/mL hygromycin B (Sigma-Aldrich, St Louis, MO). All cells were routinely tested for mycoplasma monthly using an in-house developed polymerase chain reaction test and appropriate primers covering 98% of all mycoplasma species.
mRNA Extraction, cDNA Preparation and Quantitative Polymerase Chain Reaction (qPCR) Gene Expression
Messenger RNA was extracted from the tissue using the Isolate II RNA kit (Bioline, Alexandria, New South Wales, Australia) as per the manufacturer’s instructions. Complementary DNA was prepared using the Tetro cDNA Synthesis Kit (Bioline, Alexandria, New South Wales, Australia), as per manufacturer’s instructions. The qPCR reaction mixture was prepared using the SybrGreen I Master Mix (Roche, North Ryde, New South Wales, Australia) as per manufacturer’s instructions. The qPCR was run using the Sybr Green protocol using a CFX384 Touch (Bio-Rad, Gladesville, New South Wales, Australia) using the following settings: 95°C for 2 minutes, then 40 cycles of 95°C for 15 seconds, and 62°C for 1 minute to measure primer melting temperature. Data were normalized the housekeeper genes GAPDH and ACTB2 with a Ct value of >35 being deemed not detected. Primers (Gene Works, Melbourne, Victoria, Australia) used for the study are described in Table 1.
Sequences of qPCR primers used in this study.
Fluorescence-Activated Cell Sorting Analysis of Surface and Total Expression
Surface receptor expression was measured using fluorescence-activated cell sorting (FACS) analysis, represented by FLAG tag expression. Briefly, 1 million cells were transferred into 1.5 mL Eppendorf tubes and centrifuged at 400 × g for 5 minutes at 4°C and the supernatant removed. All subsequent steps were performed on ice. The resulting pellet was resuspended in 100 μL of blocking buffer (0.01% sodium azide, PBS, 5% bovine serum albumin (BSA), 2 mM EDTA) and incubated on ice for 30 minutes. After this incubation, 100 μL of 10 μg/mL anti-FLAG tag Alexa Fluor-488 (AF-488) conjugated monoclonal antibody (Invitrogen, Waltham, MA) prepared in blocking buffer was added. The samples were incubated for 1 more hour on ice. To account for cell viability 0.5 uL of 1 mg/mL propidium iodide was then added to each sample and incubated for another 10 minutes. After the incubation, the samples were centrifuged at 400 × g for 5 minutes at 4°C and the supernatant aspirated. The samples were washed once with 100 μL wash buffer (0.04% sodium azide, PBS, 0.5% BSA, 2 mM EDTA) and resuspended. The plates were centrifuged at 400 × g for 5 minutes at 4°C once more and the supernatant aspirated. The final sample was resuspended in 200 μL wash buffer prior to analysis using a FACS Canto II flow cytometer (BD Biosciences, North Ryde, New South Wales, Australia). Settings used were as follows: AF-488 was detected by a blue laser (488 nm) with 530/30 in voltage of 310. Front scatter, side scatter, and propidium iodide voltages of 250, 450, and 380 were applied without compensation. Total receptor expression was measured using the same method described above but with the addition of 0.5% TWEEN 20 (Sigma-Aldrich, St Louis, MO) in the blocking and wash buffers.
cAMP Accumulation
The accumulation of cAMP was measured using a LANCE ULTRA cAMP Assay Kit (Perkin Elmer, Waltham, MA). Briefly, cells were seeded into transparent 384-well plates at 5000 cells per well in 25 μL stimulation buffer (Hanks’ balanced salt solution, 5 mM HEPES, 0.1% BSA and 20 μM rolipram, pH 7.4) at 37°C with 5% CO2 for 20 minutes. Compounds were serial-diluted in stimulation buffer directly from stock solutions to 7.4× the final assay concentration indicated in results. Cells were stimulated with 5 μL of each agonist for 10 minutes. cAMP accumulation was stimulated for 20 minutes by addition of either 5 μL FSK (5 μM final assay concentration for Gi-mediated response) or stimulation buffer (for Gs-mediated response). cAMP levels were detected by the addition of 6.25 μL Eu-CAMP and 6.25 μL ULight, prediluted in detection buffer (50nM HEPES, 10nM calcium chloride (CaCl2), 0.35% Triton X-100) at 1:50 and 1:150, respectively, and incubated for 60 minutes at room temperature (RT) in the dark. Fluorescence was measured using an Envision plate reader (Perkin Elmer, Waltham, MA). In all assays, a standard curve was generated to allow quantification of cAMP.
Extracelluar Signal-related Kinase (ERK) 1/2 Phosphorylation Assay
Phosphorylated extracellular signal-related kinase 1/2 (pERK1/2) was measured using AlphaScreen SureFire assay kits (Perkin Elmer, Waltham, MA). Cells were seeded into transparent poly-D-lysine (PDL)-coated (50 μg/mL; Sigma-Aldrich, St Louis, MO) 96-well plates at 40,000 cells per well and incubated overnight in 90 μL serum-free DMEM (Thermo Fisher Scientific, Waltham, MA) at 37°C/5% CO2. Compounds were serial-diluted in serum-free DMEM from stock at 10× final concentration. All compounds were profiled in the pERK1/2 assay using time-to-peak as agonist exposure time (generally 5 minutes; data not shown). Responses were terminated by removal of drugs and addition of 100 μL SureFire lysis buffer (TGR Biosciences, Thebarton, South Australia, Australia), followed by agitation on a plate shaker for 5 minutes. 5 μL cell lysate was then added to a white ProxiPlate (Perkin Elmer, Waltham, MA) followed by 8 μL of 100:600:3:3 (v−1v−1v−1v−1) SureFire activation buffer/SureFire reaction buffer/AlphaScreen donor/acceptor beads. All plates were secured with a top seal and incubated for 60 minutes at 37°C in 5% CO2 in the dark. Fluorescence was measured using an Envision plate reader (Perkin Elmer, Waltham, MA). FBS (10% v−1v−1) was used as a positive control.
GTPγ35S Binding
Cells were grown to 80%–90% confluence in 500cm2 cell-culture trays at 37°C in 5% CO2. All subsequent steps were conducted at 4°C to avoid receptor degradation. The cell-culture medium was removed and replaced with ice-cold buffer (10 mL per tray; 10 mM HEPES, 0.9% w/v sodium chloride (NaCl), 0.2% w/v EDTA, pH 7.4). Cells were scraped from the trays into a 50 mL Corning tube and centrifuged at 250 × g for 5 minutes. The supernatant was aspirated and 10 mL per 500 cm2 tray of wash buffer (10 mM HEPES, 10 mM EDTA, pH 7.4) added. Pellets were prepared after homogenization (Werker, ultra-turrax, position 6, 4 × 5 second bursts) and centrifugation at 48,000 × g at 4°C (Beckman Avanti J-251 Ultracentrifuge) for 30 minutes. The supernatant was discarded and the pellet rehomogenized in wash buffer and recentrifuged. The final pellet was suspended in ice-cold assay buffer (10 mM HEPES, 0.1 mM EDTA, pH 7.4) at a concentration of 1–2 mg/mL. Protein concentration was determined by a bicinchoninic acid assay, using BSA as a standard and aliquots maintained at −80°C until required. Compounds were prepared at 100× concentration in 100% DMSO and 2.5 μL added to each well in a 96-well assay plate together with 200 μL of assay buffer (20 mM HEPES, 10 mM MgCl2, 100 mM NaCl, 1 mM EDTA, pH 7.4), with 0.1% BSA and 30 μg per mL saponin (added fresh on the day of experimentation), containing 50 μg per mL membranes. 3.7 μM guanosine 5‘-diphosphate sodium salt was added, and the plates incubated at room temperature with gentle agitation for 30 minutes. 50 μL of GTPγ35S at a final concentration of 300 pM was then added to each well and incubated at RT with gentle agitation for 40 minutes. Bound and free GTPγ35S were separated by rapid vacuum filtration using a FilterMate Cell Harvester (Perkin Elmer, Waltham, MA) and 96-well GF/C filter plates and rapidly washed three times with ice-cold wash buffer (50 mM Tris-HCL, 10 mM MgCl2, 100 mM NaCl, pH 7.6). After drying (>4 hours), 40 μL of Microscint 20 (Perkin Elmer, Waltham, MA) was added to each well and the radioactivity quantified using single photon counting on a MicroBeta2 microplate scintillation counter (Perkin Elmer, Waltham, MA).
Calcium Assays
Assays were performed in isotonic buffer [146 mM NaCl, 5 mM potassium chloride (KCl), 1.3 mM CaCl2, 1 mM magnesium sulphate (MgSO4), 1.5 mM sodium bicarbonate (NaHCO3), 10 mM D-glucose, 10 mM HEPES, 2.5 mM probenecid, and 0.5% BSA, pH 7.4]. Assay buffer stock was prepared without probenecid or BSA, sterile filtered, and stored at 4°C until required. For assay, 100 mL of buffer was prepared by adding 0.625 mL of 400 mM probenecid [dissolved in 1 M sodium hydroxide (NaOH)] to 90 mL stock buffer, and pH adjusted to 7.4, before being made to the final volume of 100 mL. BSA (0.5% w/v) was added, and buffer used within 24 hours.
Cells were seeded into PDL-coated 96-well plates in growth medium at a density of 50,000 cells per well. The next day, plates containing a confluent monolayer of cells were washed once with 200 μL assay buffer, before being loaded with 1 μM Fluo-8-AM in 90 μL assay buffer (Abcam, Cambridge, United Kingdom) and incubated 1 hour at 37°C. Compounds were serial-diluted from stock at 10× concentrations and added on a FlexStation 3 microplate reader (Molecular Devices; Sunnyvale, CA). Fluorescence was detected using 485 nm excitation and 525 nm emission filters. Calcium influx responses were measured by the peak fluorescence post ligand addition minus the average basal fluorescence measured. All assays were performed in duplicate, and data normalized to the response of 100 μM ATP.
β-Arrestin2 Recruitment
β-arrestin2 recruitment assays were performed using Bioluminescence Resonance Energy Transfer (BRET) technology, with Rluc8-fused receptor donors and a Venus-fused β-arrestin2 acceptor (Savage et al., 2013). Bicistronic plasmids containing both receptor donor and β-arrestin2 acceptor for all six receptors of interest (CD8a-FLAG-‘GPCR’-Rluc8-IRES-βArr2-Venus) were generated for transfection into HEK293 parental cells, a method that guarantees coexpression of both BRET acceptor and donor within the same cells. This bicistronic vector system generates 4- to 7-fold higher expression of the downstream cistron (Bochkov and Palmenberg, 2006), therefore producing β-arrestin2 acceptor in excess of receptor donor within each coexpressing cell. HEK293 cells were plated at 500,000 cells per well in 6-well plates in normal growth medium and incubated overnight at 37°C in 5% CO2. Transfection was performed using a polyethylenimine (PEI) transfection protocol. For each well, 2 μg DNA of each receptor/β-arrestin2 bicistronic plasmid were diluted in 125mM NaCl to a final volume of 125μL and combined with 125 μL of 125 mM NaCl containing 15μg PEI and incubated at RT for 5 minutes. PEI/plasmid mix was added to each well and incubated for 24 hours at 37°C in 5% CO2.
Transfected cells were collected by pipetting and medium removed by centrifugation before being resuspended in phenol red-free DMEM (Thermo Fisher Scientific, Waltham, MA) with 5% FBS (3 mL media per well collected). Cells were plated into PDL-coated 96-well white CulturPlates (Perkin Elmer, Waltham, MA) at 50 μL per well. Cells were allowed to adhere overnight. Medium was removed and replaced with 80 μL serum-free phenol red-free DMEM for 1 hour (37°C), with 10 μL Coelenterazine h (100 μM; NanoLight Technology, Pinebox, AZ) added after 55 minutes, prior to reading BRET1 (480 and 530 nm emission) signals on the PheraStar (BMG Labtech, Mornington, Victoria, Australia). After three consistent basal reads, 10 μL of compounds were administered in phenol red-free DMEM (Thermo Fisher Scientific, Waltham, MA) and the plate read for a further 11 reads. Each plate contained control wells, where DMEM with no compound was added.
Compounds and Reagents
All compounds were supplied from Cayman Chemical (Ann Arbor, MI) with the exceptions of serum amyloid A (SAA) and fMLP (Sigma Aldrich, St Louis, MO), chemerin (Enzo Life Sciences, Redfern, New South Wales, Australia), and WYKMVm (Tocris Bioscience, Bristol, United Kingdom). N-(4-chlorophenyl)-N′-[2,3-dihydro-1-methyl-5-(1-methylethyl)-3-oxo-2-phenyl-1H-pyrazol-4-yl]-urea (Compound 43) was a generous gift from Dr. Cheng Xue Qin (Monash University, Melbourne, Victoria, Australia). ACT-389949 was synthesized by Servier, Suresnes, France. All SPM ligands were serially diluted directly from stock (or intermediate, where noted in Table 2) to concentrations indicated in results for each assay in either assay buffer (cAMP accumulation; Hanks’ balanced salt solution, 5 mM HEPES, 0.1% BSA and 20 μM rolipram, pH 7.4; intracellular calcium ([Ca2+]i) elevation; 146 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1 mM MgSO4, 1.5mM NaHCO3, 10 mM D-glucose, 10 mM HEPES, 2.5 mM probenecid and 0.5% BSA, pH 7.4, and GTPγ35S binding; 100% DMSO) or assay media (ERK1/2 phosphorylation assay; serum-free DMEM (cat #11995-065, Thermo Fisher Scientific, Waltham, MA) and β-arrestin2 recruitment; serum-free, phenol red-free DMEM supplemented with 1mM sodium pyruvate. Radioligand was supplied by PerkinElmer (Waltham, MA). Cell culture and molecular biology reagents were supplied by Life Technologies (Melbourne, Australia) or Thermo Fisher Scientific (Waltham, MA). Radioligand was supplied by Perkin Elmer (Waltham, MA). Cell culture and molecular biology reagents were supplied by Life Technologies (Melbourne, Victoria, Australia).
Details of compounds used in this study.
Assay specific compound dilutions and solvents used are described in the Materials and Methods and Compounds and Reagents sections.
Data Analysis.
All experiments were analyzed by nonlinear regression using Prism 9.0 (GraphPad Software Inc., San Diego, CA). All values reported in the tables are mean + S.E.M. for the indicated number of experiments. For the single concentration analysis, data are presented as the mean + S.D. normalized to each individual assay control for three to five different assay formats. The full concentration response curve data are presented as normalized to the chosen reference ligand for that receptor; e.g., FPR2/ALX; WKYMVm, BLT1; LTB4, and chemerin1; chemerin and are shown as mean + S.E.M.
qPCR data were obtained by normalizing the raw expression of genes against two housekeeping genes GAPDH and ACTB2 and calibrator samples, where ΔΔCT values using the equation as below on Bio-Rad CFX Manager software (Bio-Rad Laboratories software, Hercules, CA). Grouped data are shown as mean + S.D.

RQ refers to the relative quantity of a sample. Ref is the reference housekeeping gene in each sample, where Ref 1 is GAPDH and Ref 2 is ACTB2. GOI is the gene of interest and n refers to the number of reference targets.
Flow cytometry data were gated against parental HEK293 cells, and the level of expression was presented as the percentage of cells positively stained for FLAG-Alexa Fluor 488 in relation to parental HEK293 or as the mean fluorescence intensity of AF488. The data were analyzed using FlowJo V10 (LLC, Ashland). Grouped data are shown as mean + S.D.
Agonist concentration-response curves were fitted to sigmoidal (variable slope) using a four-parameter logistic equation:

where Y is the response measured, Top and Bottom denote the maximal and minimal asymptotes, respectively, and nH represents the Hill slope (constrained to unity where appropriate).
β-arrestin2 recruitment data were analyzed in MARS Data Analysis Software (BMG Labtech, Ortenburg, Germany) to determine BRET ratio (480 nm/530 nm), ΔBRET ratios were calculated by subtracting average basal preaddition signal (to account for minor plating/signal differences), and ΔΔBRET ratios were calculated by further subtracting average control curves (to account for signal drift over time), before analyzing the area under the curve of the subsequent time-resolved ΔΔBRET.
Ligand bias at FPR2/ALX was analyzed using the method of relative transduction ratios as a measure of ligand bias (Kenakin and Christopoulos, 2013; Riddy et al., 2017). Log(τ/KA) values, which consider both relative potency and maximal response at each pathway, were normalized to ΔLog(τ/KA) with respect to the Log(τ/KA) value for WKYMVm in each pathway, and further normalized (ΔΔLog(τ/KA)) to cAMP inhibition as the reference pathway (Riddy et al., 2017) using Microsoft Excel (2010); bias plots were generated using the radar plot feature. Where ACT-389949 displayed no response at extracelluar signal-related kinase (ERK) phosphorylation, it was given the arbitrary minimal value of 10ΔΔLog(τ/KA) = 0.001 for graphing purposes. fMLP was excluded from this analysis due to the incomplete definition of the concentration-response curves.
Results
The stable expression of FPR2/ALX, LTB4R (BLT1) and CMKLR1 (chemerin1) in HEK293 cells was confirmed by assessing mRNA and protein expression levels. FPR2/ALX, LTB4R, and CMKLR1 had similar relative mRNA expression as measured by ΔΔCT (Fig. 1A). As each receptor was tagged with an N-terminal FLAG epitope, cell-surface protein expression could be assessed by flow cytometry. Percentage of cell surface expression, as shown in Fig. 1B and quantified in Fig. 1C, revealed that FPR2/ALX and LTB4R were both highly expressed, whereas CMKLR1 was more modestly expressed at the cell surface. Using mean fluorescence intensity as a readout of expression within a cell, LTB4R was more abundantly expressed than both FPR2/ALX and CMKLR1 (Fig. 1D). Total expression levels revealed a similar pattern (Supplemental Fig. 1, A and B). After successfully demonstrating suitable levels of expression, these cell lines were next assessed for their pharmacological responses.
Expression of FPR2/ALX, LTB4R, and CMKLR1 in recombinant receptor expressing HEK293-Flp-In-CD8a-FLAG cell lines. (A) Relative expression of the three receptors. Grouped data are shown as individual data points and mean + S.D. (n = 4). (B) Representative of flow cytometry gating strategy for measurement of surface protein expression. Surface protein expression presented as (C) percentage of the total cell population positively stained with FLAG-Alexa Fluor (AF) 488 relative to the parental HEK293 cells or (D) the mean fluorescence intensity of AF488. Grouped data are shown as individual data points and mean + S.D. (n = 6).
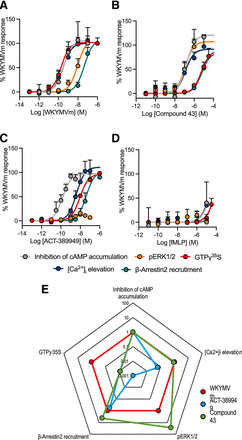
Of the 15 compounds profiled across the different signaling assays for FPR2/ALX, only four ligands displayed an appreciable response (Fig. 2, A–D; Table 3; Supplemental Fig. 2). As the peptide WKYMVm behaved as a full agonist in each assay, all data were normalized to this reference control. Robust responses were obtained to synthetic peptides (WKYMVm; Fig. 2A) and small molecules (Compound 43 and ACT-389949; Fig. 2, B and C). WKYMVm produced a rank order of potencies of GTPγ35S binding = inhibition of cAMP = [Ca2+]i elevation ≫ pERK1/2 > β-arrestin2 recruitment (Fig. 2A; Table 3; Supplemental Fig. 2, A–E). Compound 43 also displayed full agonism at FPR2, but with a different rank order of potencies: inhibition of cAMP = pERK1/2 = [Ca2+]i elevation ≫ GTPγ35S binding = β-arrestin2 recruitment (Fig. 2B; Table 3; Supplemental Fig. 2, A–E).
Concentration-response curves of the FPR2/AKX agonists (A) WKYMVm, (B) Compound 43, (C) ACT-389949, and (D) fMLP in HEK293-Flp-In-CD8a-FLAG-FPR2/ALX cells across five signaling pathways where all ligand responses are normalized to the maximal effect of WKYMVm. Grouped data are shown as mean + S.E.M. (n = 3–9). (E) Data from (A–C) were used to construct a web of bias for responses of ACT-389949 and Compound 43 relative to the reference agonist WKYMVm and normalized to the reference pathway inhibition of cAMP accumulation [axis represents 10ΔΔLog(τ/KA)]·
Potency (pEC50) and Emax (%) values for the agonist activity of the different ligands at FPR2/ALX in HEK293-Flp-In-CD8a-FLAG-‘GPCR’ stably expressing cell lines across six signaling assays.
Data represent the means ± S.E.M. Where n = 2, this represents ligands tested at a single concentration in duplicate experiments.
ACT-389949, a clinically tested FPR2/ALX small molecule agonist, produced a response in each assay for FPR2/ALX with a rank order of potency of inhibition of cAMP ≫ [Ca2+]i elevation > GTPγ35S binding > β-arrestin2 recruitment ≫ pERK1/2 (Fig. 2C; Table 3; Supplemental Fig. 2, A–E). Interestingly, its response in the pERK1/2 assay was severely reduced compared with the other pathways tested (Fig. 2C; Supplemental Fig. 2C). A time course was also performed, but no response was observed over 1 hour (data not shown).
Of all the putative endogenous ligands profiled at FPR2/ALX, only the nonselective peptide, fMLP, produced measurable responses for cAMP inhibition, GTPγ35S binding, and calcium elevation at the highest concentrations tested (Fig. 2D; Table 3; Supplemental Fig. 2). No compound produced an increase in cAMP accumulation, suggesting no apparent Gs coupling (Table 3; Supplemental Fig. 2B).
In the single concentration assay, RvE1, NPD1, and, to a lesser extent. LXA4, produced [Ca2+]i elevation responses. However, extension to full concentration response curves failed to produce any meaningful potency estimates (Table 3; Supplemental Fig. 2D).
Given the failure of ACT-389949 to stimulate pERK1/2, we quantified the extent of the signaling bias for the two small molecule FPR2 agonists. Biased agonism implies that different ligands may induce distinct signaling profiles at the same receptor leading to individual cellular outcomes (Kenakin and Christopoulos, 2013; Wootten et al., 2018). Using WKYMVm as a reference agonist and the inhibition of cAMP assay as the reference pathway, ΔΔLog(τ/KA) values (Fig. 2E) show that Compound 43 has strong bias toward pERK1/2 and β-arrestin2 recruitment but is markedly biased away from GTPγ35S binding. In addition, ACT-389949 was clearly biased away from both GTPγ35S binding and [Ca2+]i elevation, whereas it was unbiased relative to WKYMVm for the recruitment of β-arrestin2.
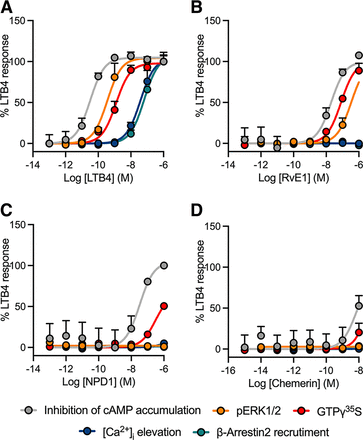
BLT1 was similarly profiled across the same set of assays. The endogenous ligand LTB4 produced full agonist responses in each assay, with rank order potencies of cAMP > pERK1/2 > GTPγ35S binding ≫ [Ca2+]i elevation = β-arrestin2 recruitment (Fig. 3A; Table 4; Supplemental Fig. 3, A–D). RvE1, an SPM ligand reported to activate multiple SPM GPCRs including BLT1, induced responses in three of the five assays with a rank order of potency of inhibition of cAMP > GTPγ35S binding > pERK1/2 (Fig. 3B; Table 4). It produced a minimal response in [Ca2+]i elevation (Supplemental Fig. 3C), which could not be robustly quantified (Table 4) and failed to activate β-arrestin2 recruitment (Fig. 3B; Table 4; Supplemental Fig. 3D).
Concentration-response curves of the BLT1 agonists (A) LTB4, (B) RvE1, (C) NPD1, and (D) chemerin in HEK293-Flp-In-CD8a-FLAG-BLT1 cells across five signaling pathways where all ligand responses are normalized to the maximal effect of LTB4. Grouped data are shown as mean + S.E.M. (n = 3–6).
Potency (pEC50) and Emax (%) values for the agonist activity of the different ligands at BLT1 in HEK293-Flp-In-CD8a-FLAG-‘GPCR’ stably expressing cell lines across six signaling assays.
Data represent the means ± S.E.M. Where n = 2 this represents ligands tested at a single concentration in duplicate experiments.
Interestingly, NPD1 was identified as a BLT1 agonist, demonstrating modest activity in the inhibition of cAMP and GTPγ35S binding assays (Fig. 3C; Table 4; Supplemental Fig. 3A), although it failed to produce a response in the remaining assays (Fig. 3C; Table 4; Supplemental Fig. 3, B–D). Similarly, chemerin, an endogenous ligand of the chemerin1 receptor, displayed weak agonist activity in the inhibition of cAMP assay (Fig. 3C; Table 4; Supplemental Fig. 3, A–D). Both RvD1 and resolvin D2 (RvD2) (1 μM) robustly inhibited FSK-induced cAMP accumulation, although neither yielded robust potency estimates in a full concentration response format (Table 4; Supplemental Fig. 2A). No signaling bias was observed for any ligand at BLT1 (data not shown).
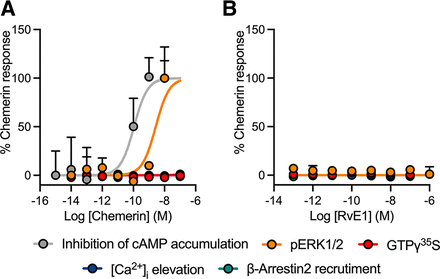
At the chemerin1 receptor, its cognate ligand, chemerin, produced a response in only the inhibition of cAMP and pERK1/2 assays (Fig. 4A; Table 5; Supplemental Fig. 4, A–D). No response was observed in the GTPγ35S binding, [Ca2+]i elevation, or β-arrestin2 recruitment assays. The reported chemerin1 agonist, RvE1, was inactive in all assays (Fig. 4B; Table 5; Supplemental Fig. 4, A–D). Unlike chemerin and RvE1, N-arachidonoyl glycine (NAGly), LTB4, and NPD1 all produced a clear response in the [Ca2+]i elevation assay when profiled at a single high concentration, although none yielded robust potency estimates in a concentration response format (Table 5; Supplemental Fig. 2C).
Concentration-response curves of the chemerin1 agonists (A) chemerin and (B) RvE1 in HEK293-Flp-In-CD8a-FLAG-chemerin1 cells across five signaling pathways where responses are normalized to the maximal chemerin effect. Grouped data are shown as mean + S.E.M. (n = 3–8).
Potency (pEC50) and Emax (%) values for the agonist activity of the different ligands at chemerin1 in HEK293-Flp-In-CD8a-FLAG-‘GPCR’ stably expressing cell lines across six signaling assays.
Data represent the means ± S.E.M. Where n = 2, this represents ligands tested at a single concentration in duplicate experiments.
Cell lines for the orphan GPCRs, GPR18, GPR32, and GPR37, were generated using the same HEK293-Flp-In-CD8a-FLAG system as described above. Stable transfection yielded modest mRNA expression for GPR18 and GPR37 (Supplemental Fig. 5A), although this expression translated into very limited surface and total receptor expression as measured by FACS (Supplemental Fig. 5, B–C). Despite a similar stable transfection regimen, there was no detectable mRNA expression of GPR32. Although the expression was extremely low, we profiled each receptor with the same set of ligands across most of the assays as described above.
Perhaps unsurprisingly, none of the ligands yielded activity in any assay (Supplemental Fig., 6 A–E), except for small responses (∼two fold over basal) for NPD1 at GPR32 (Supplemental Fig. 7B) and LXA4 at GPR37 (Supplemental Fig. 8B) in the cAMP accumulation assay (Gs). Further expression systems for GPR18 (tetracycline inducible HEK293-FlpIn-TREx and transient expression in HEK293 cells using a bicistronic vector containing both the receptor and β-arrestin2) failed to generate any functional responses (data not shown).
Discussion
Although in recent years the field of resolution pharmacology has gathered interest and ligand pairings of SPM GPCRs have been extensively investigated, there have been many inconsistencies in these findings. The role of receptor poly-pharmacology and ligand promiscuity adds extra complexities to this field (Park et al., 2020). To complement published findings, we carried out a comprehensive and methodical pharmacological analysis of six SPM GPCRs, using putative endogenous and synthetic ligands across multiple signaling assays.
FPR2/ALX has been paired with multiple proinflammatory and proresolving ligands, including LXA4, RvD1, AnxA1, and SAA (He et al., 2003; Ernst et al., 2004; Krishnamoorthy et al., 2010; Maderna et al., 2010). We found, in contrast to previous findings, that none of these ligands showed any activity in FPR2 assays, despite a consistent method of ligand handling. Interaction of these ligands with FPR2/ALX has mostly been described in radioligand binding studies, which can reflect selectivity of ligand and competitor, thus producing misleading results, and in knockout mouse models (Fiore et al., 1992; Dufton et al., 2010; Krishnamoorthy et al., 2012; Norling et al., 2012) with limited data from functional studies.
The proresolving effects of LXA4, including stimulation of efferocytosis, have been suggested to be mediated by FPR2/ALX activation (Maderna et al., 2010). However, other studies questioned these effects with LXA4 unable to produce functional responses in [Ca2+]i elevation and β-arrestin2 recruitment assays in FPR2-expressing cells (Christophe et al., 2002; Forsman et al., 2011). RvD1 induced β-arrestin2 recruitment at subnanomolar potency in the DiscoveRx Pathhunter system when FPR2/ALX was expressed in HEK293 or CHO cell backgrounds (Krishnamoorthy et al., 2010). This result was somewhat surprising, as typically β-arrestin recruitment potencies from this assay format are close to their binding affinity (Riddy et al., 2012); this result would suggest other RvD1 functional potencies in the picomolar range.
However, using BRET rather than enzyme complementation, no effect of RvD1 (or any other SPM) was detected in the β-arrestin2 recruitment assay, highlighting clear inconsistencies within the field. AnxA1, a glucocorticoid-mediator protein, also showed no activity in any assay, again at odds with previous western blot analysis of AnxA1-activated ERK1/2 in FPR2/ALX transfected HEK293 cells (Hayhoe et al., 2006).
LXA4, RvD1, and AnxA1 exert proresolving physiologic effects, including inhibition of neutrophil infiltration, enhancement of apoptosis and efferocytosis of apoptotic neutrophils, and downregulation of proinflammatory mediators (El Kebir et al., 2009; Norling et al., 2012; Lee and Surh, 2013). These actions are putatively linked to FPR2, so their lack of activity in our signaling assays is not clear. It may be possible that in a native context additional receptors or scaffolding proteins (absent in HEK293 cells) are required for these physiologic effects, or due to limitations in compound availability and solubility, their effects may only be observed at concentrations higher than those tested within this study.
The SPM ligands, particularly the putative endogenous lipoxins and resolvins, are notoriously labile and prone to issues associated with handling, storage, and preparation. However, wherever possible all ligands were prepared and diluted as recommended by manufacturers and consistent with best practice in the field. For example, we observed robust signaling with RvE1 at BLT1 indicating that ligand preparation is an unlikely explanation for its lack of activity at other receptors.
The synthetic FPR2/ALX ligands, WKYMVm and Compound 43, induced robust responses across all FPR2/ALX signaling assays, corroborating published data (Zhou et al., 2007; Forsman et al., 2011; Qin et al., 2017). However, in our hands, the first-in-class investigational agonist, ACT-389949, failed to induce ERK1/2 phosphorylation but generated a robust response at other pathways. ACT-389949 progressed to phase I clinical trials but was discontinued due to lack of efficacy on neutrophil recruitment in a lipopolysaccharide challenge model in healthy human subjects (Stalder et al., 2017). Follow-up pharmacological studies revealed Gq-independent transient rises in intracellular Ca2+ and recruitment of β-arrestin (Lind et al., 2019), which are further confirmed by our findings.
Activation of the MEK/ERK pathway is crucial for the proinflammatory function of neutrophils, including phagocytosis, delayed apoptosis and activation of NADPH oxidase to form neutrophil extracellular trap, which helps destroy microbes (Hakkim et al., 2011). Conversely, inhibition of MEK-dependent ERK activation significantly impairs lipopolysaccharide-stimulated neutrophil mediated release of proinflammatory cytokines [C-X-C motif chemokine ligand 8 (CXCL8), C-C motif chemokine ligand 3 (CCL3), CCL4] (Simard et al., 2015), which are essential for further recruitment of neutrophils and other phagocytic immune cells (Tecchio and Cassatella, 2016). ERK1/2 activation is also linked to host immune responses, including macrophage activation (Willmann et al., 2020), uptake of mycobacteria (Li et al., 2015), and key effector functions of CD8+ T cells (Crawford et al., 2013). ERK1/2 signaling is implicated in multiple inflammatory diseases, including autoimmune disorders. Defective ERK1/2 signaling is hypothesized to contribute to lupus pathogenesis, causing gene dysregulation and autoreactivity in T cells (Gorelik and Richardson, 2010), and there is T cell-ERK1/2 phosphorylation impairment in patients with active lupus (Deng et al., 2001).
ACT-389949s failure to induce ERK1/2 phosphorylation in HEK293 cells may provide further evidence to explain its limited clinical efficacy, in addition to the observation that it stimulates robust receptor internalization. Collectively, these effects may need to be considered when developing new synthetic FPR2/ALX small molecule agonists with greater clinical efficacy.
We confirmed previous reports that RvE1 is a BLT1 agonist, with potencies in the mid–high nanomolar range across different assays. However, previous studies identified RvE1 in serum at low picomolar concentrations (Lotfi et al., 2020), perhaps questioning the physiological role of this ligand at BLT1. We also verified robust chemerin activity at chemerin1 in both cAMP inhibition and ERK1/2 phosphorylation assays. However, the ligand showed no activity in GTPγ35S binding, [Ca2+]i elevation, or β-arrestin2 recruitment assays. These data contrast previous studies (Wittamer et al., 2003; Luangsay et al., 2009), which showed GTPγ35S binding and [Ca2+]i elevation to chemerin in chemerin1-CHO-K1 cells and mouse bone marrow-derived dendritic cells and peritoneal macrophages. This further underpins the critical role of cell background in studies of chemerin1 function. In addition, it should be noted that differences in assay kinetics, incubation time, and temperature may further influence these results (Klein Herenbrink et al., 2016).
A significant finding of the study was the identification of new ligand-receptor pairings, with chemerin at concentrations >10 nM and NPD1 at nanomolar concentrations activating BLT1 in cAMP and GTPγ35S binding assays.
NPD1 is predominantly found in the brain and is known to have neuroprotective actions (Bazan, 2007). It was recently paired with the orphan receptor, GPR37 (Bang et al., 2018; Bang et al., 2021). As a potential additional receptor for NPD1, BLT1 is also broadly expressed in the human brain, notably in neurons and glial cells of the hippocampus, entorhinal cortex, basal forebrain, cingulate gyrus, Brodmann area 46, and cerebellum (Emre et al., 2020). This wide-ranging expression, which includes immune cells, suggests that BLT1 may play a role in the neuroprotective actions of NPD1.
Conversely, this study failed to replicate some previously reported ligand-receptor pairings. For example, RvE1 did not induce a response when profiled at chemerin1. Other studies have also failed to show an effect of RvE1 on chemerin1, including attenuation of PMN migration in chemerin1−/− cells (Arita et al., 2005) and displacement of tritiated-RvE1 ([3H]RvE1) by LTB4, but not chemerin on human neutrophils (Arita et al., 2007). Activation of BLT1 but not chemerin1 within our study supports these original findings and may suggest that the proresolving effects of RvE1 on neutrophil function are due to BLT1 activation.
Despite our broader aim to examine the pharmacological profile of six GPCRs, the orphans, GPR18, GPR37, and GPR32, proved extremely difficult to express, as measured at mRNA or protein level. It is difficult to ascertain why these orphan receptors failed to express. All six GPCRs were generated using the same HEK293-Flp-In-CD8a-FLAG cell system. It is unlikely, but possible, that the FLAG epitope impairs expression, although it is commonly used for measuring GPCR expression (Beerepoot et al., 2013). GPR18 has been expressed in other recombinant studies, which suggests that it has high constitutive activity (Finlay et al., 2016). If so, this could hinder prolonged cell surface location in a nominally stable expression system, although other attempts at transient expression in our hands yielded similar results. Relatively little is known about GPR32 or GPR37, but a similar mechanism may explain poor expression; alternatively, it could reflect the cell background.
In summary, a systematic, multipathway evaluation of a range of putative endogenous and synthetic SPM ligands at six GPCRs has revealed two new ligands for BLT1 and confirmed some previous pairings. However, in several cases, notably for FPR2, we were not able to replicate previous studies. It may be that different cell backgrounds (e.g., CHO, THP-1, or HL-60s) or deeper signaling analyses (e.g., TruPath G protein-coupling) (Olsen et al., 2020) are required to fully establish the detailed pharmacology of some of these putative endogenous ligands. However, we did reveal marked signaling bias downstream of FPR2 for synthetic agonists, which may further help guide the design of FPR2/ALX agonists for the treatment of multiple inflammatory diseases, including autoimmune disorders.
Authorship Contributions
Participated in research design: Merlin, Park, Allinne, Langmead, Riddy.
Conducted experiments: Merlin, Park, Vandekolk, Riddy.
Contributed new reagents or analytic tools: Fabb.
Performed data analysis: Merlin, Park, Vandekolk, Riddy.
Wrote or contributed to the writing of the manuscript: Merlin, Park, Summers, Langmead, Riddy.
Footnotes
- Received October 7, 2021.
- Accepted January 25, 2022.
This work received no external funding but was partially funded by Servier Laboratories.
No author has an actual or perceived conflict of interest with the contents of this article.
↵1 J.M. and J.P. contributed equally to this work.
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- ACT-389949
- N-(2-{[4-(1, 1-difluoroethyl)-1, 3-oxazol-2-yl]methyl}-2H-1, 2, 3-triazol-4-yl)-2-methyl-5-(3-methylphenyl)-1, 3-oxazole-4-carboxamide
- ALX
- lipoxin A4 receptor
- AnxA1
- annexin A1
- BLT1
- leukotriene B4 receptor 1
- BRET
- Bioluminescence Resonance Energy Transfer
- BSA
- bovine serum albumin
- [Ca2+]i
- intracellular calcium elevation; Compound 43, N-(4-chlorophenyl)-N′-[2, 3-dihydro-1-methyl-5-(1-methylethyl)-3-oxo-2-phenyl-1H-pyrazol-4-yl]-urea
- DMEM
- Dulbecco’s modified eagle’s medium
- FACS
- fluorescence-activated cell sorting
- fMLP
- N-formylmethionine-leucyl-phenylalanine
- FPR2
- formyl peptide receptor 2
- GPCR
- G protein-coupled receptor
- GTPγ35S
- [35S]-guanosine 5′-(γ-thio)triphosphate
- HEK
- human embryonic kidney
- LTB4
- leukotriene B4
- LXA4
- lipoxin A4
- NAGly
- N-arachidonoyl glycine
- NPD1
- neuroprotectin D1
- PDL
- poly-D-lysine
- PEI
- polyethylenimine
- pERK
- phosphorylated extracellular signal-related kinase
- qPCR
- quantitative polymerase chain reaction
- RT
- room temperature
- RvD1
- resolvin D1
- RvD2
- resolvin D2
- RvE1
- resolvin E1
- SAA
- serum amyloid A
- SPM
- specialized proresolving mediators
- Copyright © 2022 by The American Society for Pharmacology and Experimental Therapeutics