Abstract
Glucocorticoids act via the glucocorticoid receptor (GR; NR3C1) to downregulate inflammatory gene expression and are effective treatments for mild to moderate asthma. However, in severe asthma and virus-induced exacerbations, glucocorticoid therapies are less efficacious, possibly due to reduced repressive ability and/or the increased expression of proinflammatory genes. In human A549 epithelial and primary human bronchial epithelial cells, toll-like receptor (TLR)-2 mRNA and protein were supra-additively induced by interleukin-1β (IL-1β) plus dexamethasone (IL-1β+Dex), interferon-γ (IFN-γ) plus dexamethasone (IFN-γ+Dex), and IL-1β plus IFN-γ plus dexamethasone (IL-1β+IFN-γ+Dex). Indeed, ∼34- to 2100-fold increases were apparent at 24 hours for IL-1β+IFN-γ+Dex, and this was greater than for any single or dual treatment. Using the A549 cell model, TLR2 induction by IL-1β+IFN-γ+Dex was antagonized by Org34517, a competitive GR antagonist. Further, when combined with IL-1β, IFN-γ, or IL-1β+IFN-γ, the enhancements by dexamethasone on TLR2 expression required GR. Likewise, inhibitor of κB kinase 2 inhibitors reduced IL-1β+IFN-γ+Dex–induced TLR2 expression, and TLR2 expression induced by IL-1β+Dex, with or without IFN-γ, required the nuclear factor (NF)-κB subunit, p65. Similarly, signal transducer and activator of transcription (STAT)-1 phosphorylation and γ-interferon-activated sequence-dependent transcription were induced by IFN-γ. These, along with IL-1β+IFN-γ+Dex–induced TLR2 expression, were inhibited by Janus kinase (JAK) inhibitors. As IL-1β+IFN-γ+Dex–induced TLR2 expression also required STAT1, this study reveals cooperation between JAK-STAT1, NF-κB, and GR to upregulate TLR2 expression. Since TLR2 agonism elicits inflammatory responses, we propose that synergies involving TLR2 may occur within the cytokine milieu present in the immunopathology of glucocorticoid-resistant disease, and this could promote glucocorticoid resistance.
SIGNIFICANCE STATEMENT This study highlights that in human pulmonary epithelial cells, glucocorticoids, when combined with the inflammatory cytokines interleukin-1β (IL-1β) and interferon-γ (IFN-γ), can synergistically induce the expression of inflammatory genes, such as TLR2. This effect involved positive combinatorial interactions between NF-κB/p65, glucocorticoid receptor, and JAK-STAT1 signaling to synergistically upregulate TLR2 expression. Thus, synergies involving glucocorticoid enhancement of TLR2 expression may occur in the immunopathology of glucocorticoid-resistant inflammatory diseases, including severe asthma.
Introduction
Synthetic glucocorticoids, administered as inhaled corticosteroids (ICS), are widely and successfully used to treat mild to moderate asthma (Rhen and Cidlowski, 2005). However, in patients with severe neutrophilic (a.k.a. T2 low) asthma or viral exacerbations or those who smoke or have chronic obstructive pulmonary disease, ICS medications exhibit reduced clinical efficacy (Barnes, 2013). Thus, partial, or complete, unresponsiveness to ICS therapy in such conditions is often referred to as glucocorticoid resistance or insensitivity. Furthermore, the elevation of ICS dose and/or the inclusion of oral glucocorticoids with the aim of achieving increased clinical effectiveness in such patients leads to systemic adverse effects that include reduced hypothalamus-pituitary-adrenal axis function, weight gain, osteoporosis, hypertension, and hyperglycemia leading to diabetes (Liu et al., 2013).
Glucocorticoids act on the glucocorticoid receptor (GR; NR3C1) to suppress the expression of inflammatory mediators and thereby attenuate inflammation (Rhen and Cidlowski, 2005; Newton and Holden, 2007). These effects of glucocorticoid on airway epithelial cells are also thought to be essential in the resolution of lung inflammation caused by allergen inhalation (Klaßen et al., 2017). Glucocorticoids diffuse through the cell membrane and bind to cytoplasmic GR to drive its nuclear translocation. In the nucleus, ligand-bound GR acts as a transcription factor by binding to palindromic glucocorticoid response elements (GREs), which are located in the regulatory regions of target genes. These DNA elements are responsible for driving glucocorticoid-induced expression of many metabolic and anti-inflammatory genes (Jantzen et al., 1987; Imai et al., 1990; Newton and Holden, 2007). However, ligand-activated GR may also interact with inflammatory transcription factors, such as nuclear factor κB (NF-κB) and AP-1, and this effect is often suggested as directly causing transcriptional repression (De Bosscher and Haegeman, 2009). Despite this, an increasing body of evidence clearly shows that GR can positively interact with inflammatory transcription factors, resulting in additive, or even synergistic, transcriptional outcomes (Hofmann and Schmitz, 2002; Altonsy et al., 2014; Miyata et al., 2015; Newton et al., 2017; Oh et al., 2017). Thus, glucocorticoid-activated mechanisms variously increase or decrease the expression of inflammatory genes. As a consequence, glucocorticoids should more accurately be considered as being immunomodulatory rather than simply being “anti-inflammatory” (Busillo and Cidlowski, 2013; Cruz-Topete and Cidlowski, 2015; Newton et al., 2017; Amrani et al., 2020).
Aside from being a primary target for therapy, the airway epithelium is first in line to encounter inhaled particulate matter and microbes that enter the airways (Tam et al., 2011; Lambrecht and Hammad, 2012). Although these structural cells serve as a physical barrier, they are also fundamental in shaping inflammatory responses (Tam et al., 2011; Lambrecht and Hammad, 2012). In response to environmental insults and stimuli, airway epithelial cells release proinflammatory cytokines, chemokines, and inflammatory enzymes via activation of inflammatory signaling pathways (Lambrecht and Hammad, 2012). Cytokines, such as interleukin-1β (IL-1β) and tumor necrosis factor (TNF)-α are central to inflammatory responses, to a large extent due to the activation of the IκB kinase (IKK)–NF-κB pathway (Pahl, 1999). Similarly, interferon-γ (IFN-γ) activates Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling and is a key regulator of immune responses related to viral infection and humoral immunity (Schroder et al., 2004). Interactions between IFN-γ- and TNF-mediated pathways are also implicated in promoting glucocorticoid resistance (Tliba et al., 2008; Britt et al., 2019). Furthermore, IL-1β and IFN-γ play a variety of roles in airway inflammatory responses, including in severe asthma and both bacterial and viral exacerbations of asthma (Ngoc et al., 2005; Lambrecht et al., 2019). Since glucocorticoids are frequently employed as a treatment in these conditions, albeit with reduced efficacy, the coregulation of inflammatory genes by glucocorticoids IL-1β and IFN-γ is likely to be important to the mechanisms that underlie glucocorticoid insensitivity.
Toll-like receptor (TLR)-2 is located on the surface of airway epithelial cells and recognizes pathogen-associated molecular patterns present in many bacteria, viruses, and other microbes (Akira et al., 2001). In a prior study (Bansal et al., 2022), we investigated the synergistic induction of TLR2 expression in airway epithelial cells by glucocorticoids and proinflammatory cytokines (IL-1β and TNF), which involved positive cooperativity between GR and NF-κB. However, although studies also suggest that glucocorticoids enhance TLR2 expression induced by combinations of TNF and IFN-γ (Homma et al., 2004; Winder et al., 2009), molecular mechanisms underlying such effects are not established. As many biologic effects of IFN-γ are manifested via the transcription factor STAT1 (Schroder et al., 2004), we interrogated possible roles and interactions between GR, NF-κB and STAT1 in the regulation of TLR2 expression.
Materials and Methods
Stimuli, Drugs, and Other Inhibitors
Recombinant human IL-1β (R&D Systems) was dissolved in PBS containing 0.1% bovine serum albumin (Sigma-Aldrich). Recombinant human IFN-γ (R&D Systems) was dissolved in deionized sterile water. Budesonide (gift from AstraZeneca, Sweden), dexamethasone (Dex; Sigma-Aldrich), Organon 34517 (Org34517) (gift from Chiesi Farmaceutici, Parma, Italy), N-(6-chloro-9h-pyrido[3,4-b]indol-8-yl)-3-pyridinecarboxamide (PS-1145) (Sigma-Aldrich), N-(6-Chloro-7-methoxy-9H-pyrido[3,4-b]indol-8-yl)-2-methyl-3-pyridinecarboxamide dihydrochloride (ML120B) (SML1174, Sigma-Aldrich), 2-[(Aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide (TPCA-1) (2559, Tocris), (S)-5-chloro-N2-(1-(5-fluoropyrimidin-2-yl)ethyl)-N4-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine (AZD1480) (5617, Tocris), βR-cyclopentyl-4-(7H-pyrrolo[2,3-day]pyrimidin-4-yl)-1H-pyrazole-1-propanenitrile (ruxolitinib) (7067, Tocris), and 3-Azetidineacetonitrile, 1-(ethylsulfonyl)-3-[4-(7H-pyrrolo[2,3-day]pyrimidin-4-yl)-1H-pyrazol-1-yl]- (barcitinib) (7222, Tocris) were dissolved in DMSO as stocks of 10 mM. Final DMSO concentrations on cells were ≤0.1%. G418 (A1720, Sigma-Aldrich) was dissolved in sterile water as stocks of 100 mg/ml.
Submersion Culture of Cells
A549 human pulmonary type II epithelial cells (American Type Culture Collection CCL-185; Research Resource Identifier: CVC_0023L) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and 2 mM L-glutamine (all Thermo Fisher Scientific). Nontransplanted normal human lungs obtained from the tissue retrieval service at the International Institute for the Advancement of Medicine (Edison, NJ) were used to isolate primary human bronchial epithelial cells (pHBECs) as previously described (Hudy et al., 2010). No personal identifying information was provided, and local ethics approval was granted via the Conjoint Health Research Ethics Board of the University of Calgary (Study ID: REB15-0336). Cells were grown at 37°C in 5% CO2:95% air as submersion cultures using complete airway epithelial cell medium (PromoCell). To arrest growth and reducing activation of pathways, cells were incubated in serum- and additive-free basal media overnight prior to all experiments. A549 cells were periodically checked for possible mycoplasma contamination and tested negative. pHBECs were not tested for mycoplasma contamination.
Air-Liquid Interface Culture of pHBECs
pHBECs were grown in air-liquid interface (ALI) culture using transwell inserts (3408, Corning) as previously described (Warner et al., 2019; Michi and Proud, 2021). Over 5 to 6 weeks, this method produces a highly differentiated cell layer that is made up of mucus-producing goblet cells, ciliated columnar epithelial, and basal cells that very closely resemble a normal airways epithelium (Michi and Proud, 2021). Details of our ALI methodology were recently published (Bansal et al., 2022). Only highly differentiated ALI cultures were used. Prior to experiments, cells were fed basally with PneumaCult-ALI basal media (05002, StemCell), with no supplements for 18 hours. The cells were then washed with PBS to remove excess mucus prior to both apical and basolateral application of drugs and/or stimuli in fresh medium. Apical treatments (200 µl/well) were diluted in 0.025 M HEPES in F12 and removed after 6 hours, whereas basolateral treatments (1 ml/well) were in PneumaCult-ALI basal medium.
RNA Extraction, cDNA Synthesis, and Quantitative Polymerase Chain Reaction
Total RNA was extracted, and 500 ng was used for cDNA synthesis as described (Bansal et al., 2022). cDNA was diluted 1:5 prior to using 2.5 µl for polymerase chain reaction (StepOnePlus, Applied Biosciences or QuantStudio3, Thermo Fisher Scientific) with Fast SYBR Green Master Mix (4385618, Thermo Fisher Scientific) and primers specific for TLR2 (forward: GCT GCT CGG CGT TCT CTC and reverse: AAG CAG TGA AAG AGC AAT GGG) and GAPDH (forward: TTC ACC ACC ATG GAG AAG GC and reverse: AGG AGG CAT TGC TGA TGA TCT) as described (Bansal et al., 2022). Relative cDNA concentrations were obtained using standard curves generated by serial dilution of a stimulated cDNA sample analyzed alongside experimental samples. Amplification conditions were 95°C for 20 seconds, then 40 cycles of 95°C for 3 seconds and 60°C for 30 seconds. Primer specificity was assessed by melt curve analysis using 95°C for 3 seconds and 60°C for 30 seconds followed by ramping to 95°C at 0.1°C/s with continuous fluorescent measurement. A single peak in the change of fluorescence with temperature was indicative of primer specificity.
Western Blotting
Western blotting was carried out using standard methods (King et al., 2009). Following cell lysis, proteins were size-fractionated in SDS-PAGE gels prior to transferring to nitrocellulose membranes. After blocking, membranes were probed with primary antibodies against TLR2 (12276, Cell Signaling; RRID: AB_2797867 and ab108998, Abcam; RRID: AB_10861644), IκBα (sc-371, SantaCruz; RRID: AB_10861644), Ser32/Ser36 phosphorylated IκBα (9246, Cell Signaling; RRID: AB_2267145), p65 (sc-8008, SantaCruz RRID: AB_628017), Ser536 phosphorylated p65 (3036, Cell Signaling; RRID: AB_331281), GR (PA1-511A, Thermo Fisher Scientific; RRID: AB_2236340), Tyr701 phosphorylated STAT1 (9167, Cell Signaling; RRID: AB_561284), Ser727 phosphorylated STAT1 (9177, Cell Signaling; RRID: AB_2197983), STAT1 (9172, Cell Signaling; RRID: AB_2198300), or GAPDH (MCA4739, Bio-Rad; RRID: AB_1720065). Following overnight incubation at 4°C, the membranes were washed in Tris-buffered saline with 0.05% v/v Tween 20 detergent and incubated, as appropriate, with 1:5000 or 1:10,000 dilutions of rabbit or mouse horseradish peroxidase–conjugated secondary immunoglobulin (Jackson ImmunoResearch) at room temperature. Immune complexes were detected by enhanced chemiluminescence (Bio-Rad), and images were captured using a ChemiDoc Touch imaging system (Bio-Rad). Densitometric analysis was performed using ImageLab software (Bio-Rad). Note: TLR2 western blots shown in Fig. 1A were generated using ab108998 (Abcam); all other TLR2 blots were generated using 12276 (Cell Signaling) due to the discontinuation of ab108998.
Combinatorial effect of IL-1β, IFN-γ, and glucocorticoid on TLR2 protein expression in pulmonary epithelial cells. (A) A549 cells or (B) pHBECs grown in submersion culture were either not treated or treated with IL-1β (1 ng/ml), IFN-γ (10 ng/ml), and/or Dex (1 µM) as indicated. Cells were harvested at 6 or 24 hours for western blot analysis of TLR2 and GAPDH. Representative blots from N independent experiments are shown. (Upper graphs) Densitometric data were normalized to GAPDH, expressed as fold of untreated, and mean ± S.D. were plotted as bar graphs with overlaid scatter plots. Significance was tested using ANOVA with a Tukey post hoc test. Asterisks (*, **, or ***) represent significance relative to untreated. Hash (#, ##, or ###) represent significance relative to IL-1β+IFN-γ+Dex. (Lower graphs) The sum of each effect (i.e., fold − 1) within a group of treatments (sum) and the effect (fold − 1) for each combination treatment (comb) is expressed as mean ± S.D. and plotted as bar graphs overlaid with scatter plots. Significance was tested by paired t test. *, #P ≤ 0.05; **, ##P ≤ 0.01; ***, ###P ≤ 0.001. ns, nonspecific band.
siRNA-Mediated Knockdown
Pools of four nontargeting short interfering RNAs (siRNAs) (SI03650325, SI03650318, SI04380467, 1022064), GR siRNAs (SI00003745, SI02654757, SI00003766, SI02654764), RELA siRNAs (SI02663101, SI05146204, SI00301672, SI02663094), and STAT1 siRNAs (SI03119025, SI04950960, SI02662884, SI05078556) (all from Qiagen) were mixed with 3 µl Lipofectamine RNAiMax (13778150, Thermo Fisher Scientific) in 100 µl Opti-MEM (31985070, Thermo Fisher Scientific) before incubation at room temperature for 5 minutes. This mixture was then added to A549 cells, at approximately 70% confluency, in serum-containing growth medium and incubated for 24–48 hours until the cells were confluent.
Luciferase Reporter Constructs and Assay
A549 cells containing the NF-κB reporter 6κBtk.luc and the GRE reporter 2×GRE.luc are previously described (Newton et al., 1998; Chivers et al., 2004). To generate the 3×STAT1.luc reporter, oligonucleotides containing three tandem repeats of a consensus STAT1 motif (JASPAR database), also known as a γ-interferon–activated sequence (GAS) (Decker et al., 1997), were designed to generate flanking KpnI and XhoI sticky ends. These oligonucleotides were annealed by heating to 95°C for 5 minutes prior to cooling to room temperature over a 45-minute period in 10 mM Tris (pH 7.5), 50 mM NaCl, and 1 mM EDTA. Annealed STAT1 oligonucleotides were cloned into KpnI (R3142, New England Biolabs) and XhoI (R0146, New England Biolabs) digested pGL3.TATA.neo vector (Catley et al., 2004) to generate pGL3.3×STAT1.TATA.neo. Twelve micrograms of each reporter, or empty vector (as control), was transfected into subconfluent A549 cells in T-75 flasks using Lipofectamine 2000 (11668019, Thermo Fisher Scientific) 24 hours prior to addition of 1 mg/ml G418 (Sigma-Aldrich). After 14–21 days, foci of G418-resistant cells were passaged in the presence of G418 (0.5 mg/ml) prior to cryo-storage. Luciferase assays were performed using the Firefly Luciferase Assay Kit 2.0 (30085, Biotium Inc.), and luminescence was measured in a 20/20n luminometer (Promega).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
Cell viability was measured using a 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Assay. MTT (#298-93-1, Sigma-Aldrich) was dissolved in Hank’s balanced salt solution (14170112, Thermo Fisher Scientific) to achieve a concentration of 1 mg/mL. Cells grown in 24-well plates were pretreated with IKK2 inhibitors 90 minutes prior to being treated with IL-1β (1 ng/mL). After 6 hours, supernatants were removed, and 100 μL of MTT solution was added to each well. After 10–20 minutes of incubation in 37°C in 5% CO2/95% air, MTT solution was removed, and 200 μL of DMSO was added. The solution was transferred to a 96-well plate, and absorbance was measured at 570 nm.
Data Presentation and Statistical Analysis
Data were assumed to be normally distributed and are generally presented as means ± S.D. with scatterplots to show individual data points. Multiple comparisons between groups of normally distributed data were made using one-way ANOVA with the appropriate post hoc test (Tukey’s or Dunnett’s) as indicated. Comparisons between two treatment groups were made using two-tailed, paired Student t tests. The number of independent experiments (N) is indicated in each graph, and sample sizes (N) for experiments were not prespecified. To test for greater than simple additivity, the sum of the effects (i.e., fold − 1) of each treatment (IL-1β, IFN-γ, or glucocorticoid alone) (sum) was compared with the effect of cotreatment (comb). GraphPad Prism software (version 6.01 and 10; Dotmatics, Boston, MA) was used for statistical analyses and graph plotting. All analyses in the current study were exploratory and were not designed to test prespecified statistical null hypotheses. Consequently, all calculated P values are descriptive and should not be viewed as hypothesis testing.
Results
Regulation of TLR2 Protein Expression by IL-1β, IFN-γ, and Glucocorticoid in Pulmonary Epithelial Cells
In previous studies, IL-1β, at 1 ng/ml, produced maximal, or near maximal, NF-κB DNA-binding activity as well as NF-κB–dependent reporter activity and cytokine release in A549 cells (Newton et al., 1996; Catley et al., 2004). Similarly, dexamethasone, at 1 μM, and budesonide (Bud), a clinically relevant glucocorticoid, at 300 nM, elicited maximal activation of simple GRE-dependent transcription and maximal expression of glucocorticoid-induced genes (Kelly et al., 2012; Rider et al., 2015). Since dexamethasone and budesonide produced near identical expression profiles in A549 cells (Mostafa et al., 2019), dexamethasone was used as the representative glucocorticoid in the A549 cells and pHBECs grown in submersion cultures, whereas budesonide was used in pHBECs grown at ALI.
In A549 cells, IL-1β at 1 ng/ml and dexamethasone at 1 μM each very modestly induced TLR2 protein at 6 hours, with slightly increased effects observed at 24 hours (Fig. 1A, upper panels). These effects were enhanced by IL-1β plus dexamethasone (IL-1β+Dex) such that the combination effect of IL-1β+Dex (i.e., fold IL-1β+Dex − 1) (comb) was greater (P ≤ 0.01) than the summation (sum) of the effects of IL-1β and dexamethasone (i.e., fold IL-1β-1 + fold dexamethasone-1) (Fig. 1A, lower panels). Since these data are consistent with our prior report in which IL-1β+Dex–induced TLR2 mRNA expression was maximal at 6 hours, and greater TLR2 protein expression was apparent at 24 hours (Bansal et al., 2022), these times were also used to investigate the additional effect of IFN-γ on IL-1β+Dex–induced TLR2. Although IFN-γ (10 ng/ml), at a concentration that induces chemokine release in epithelial cells (Tudhope et al., 2007), produced very little effect at 6 hours, cotreatment with IL-1β produced a modest increase in TLR2 protein expression. However, cotreatment of IFN-γ plus dexamethasone (IFN-γ+Dex) increased TLR2 protein expression in a manner that was greater (P ≤ 0.05) than simple additivity (Fig. 1A). Thus, by 24 hours, IFN-γ+Dex increased TLR2 protein levels by 10.7-fold and again revealed supra-additivity (P ≤ 0.05). Upon combined stimulation with IL-1β plus IFN-γ plus dexamethasone (IL-1β+IFN-γ+Dex), TLR2 protein was increased (P ≤ 0.05) to 11.3-fold at 6 hours, and this was further increased to 33.6-fold (P ≤ 0.05) by 24 hours (Fig. 1A). In each case, this response was markedly (P ≤ 0.05) more than each individual treatment, and, at 24 hours, TLR2 protein expression induced by IL-1β+IFN-γ+Dex was significantly greater (P ≤ 0.01) than that for each single treatment (Fig. 1A, upper panels;) as well as consistently and significantly (all P ≤ 0.05) more than for each of the three dual treatments (Supplemental Fig. 1A). Moreover, comparison of the sum of the effects of each stimulus with the effect of the combined stimulation revealed obvious (P ≤ 0.05 and 0.01, respectively) supra-additivity between IL-1β, IFN-γ, and dexamethasone at both 6 and 24 hours (Fig. 1A, lower panels).
In pHBECs grown in submersion culture, IL-1β, IFN-γ, and dexamethasone alone produced low levels of TLR2 protein, whereas treatment with IL-1β+Dex, IL-1β+IFN-γ, or IFN-γ+Dex each more clearly induced TLR2 protein at 24 hours (Fig. 1B, upper graph). Summation of the effects for each of these pairs of stimuli (sum) was less (P ≤ 0.05) than the respective combination effect (comb), and therefore supra-additivity is revealed (Fig. 1B, lower graph). In the case of treatment with IL-1β+IFN-γ+Dex, TLR2 protein was increased by ∼2100-fold (P ≤ 0.05), and this was to a higher level than for each of IL-1β, IFN-γ, or dexamethasone alone (Fig. 1B, upper graph). Comparing the additive effects for each of IL-1β, IFN-γ, and dexamethasone (sum) with the combination effect of IL-1β+IFN-γ+Dex (comb) showed marked (P ≤ 0.05) supra-additivity between these stimuli on TLR2 protein (Fig. 1B, lower graph).
In pHBECs grown at ALI, whereas each of IL-1β, IFN-γ, and budesonide produced little increase in TLR2 protein at both 6 and 24 hours, induction (all P ≤ 0.01) was produced by IL-1β plus budesonide (IL-1β+Bud) and IL-1β plus IFN-γ plus budesonide (IL-1β+IFN-γ+Bud) (Supplemental Fig. 2A, upper panels). However, in ALI cultures of pHBECs, none of the combinations of IL-1β, IFN-γ, and budesonide revealed statistically significant supra-additivity on TLR2 protein at either 6 or 24 hours (Supplemental Fig. 2A, lower panels). Nevertheless, in the case of IL-1β+Bud and IL-1β+IFN-γ+Bud, trends toward a greater effect for the combination treatment (comb) compared with the sum of the effects of each stimulus (sum) were apparent.
Effect of IL-1β, IFN-γ, and Glucocorticoid on TLR2 mRNA Expression in Pulmonary Epithelial Cells
TLR2 mRNA was only marginally induced by IL-1β, IFN-γ, or dexamethasone alone at both 6 and 24 hours in A549 cells (Fig. 2A, upper panels). However, the combination of IL-1β+Dex increased TLR2 mRNA at 6 hours in a manner that was greater (P ≤ 0.01) than simple additivity (Fig. 2A). Similarly, dexamethasone also enhanced IFN-γ–induced TLR2 mRNA at 6 and 24 hours such that the effect of cotreatment with IFN-γ+Dex (comb) was higher (P ≤ 0.05) than the sum of the effects of IFN-γ and dexamethasone alone (sum) (Fig. 2A). These data indicate supra-additivity between IFN-γ and dexamethasone on TLR2 mRNA at both 6 and 24 hours. Cotreating A549 cells with IL-1β+IFN-γ also led to slight increases in TLR2 mRNA at 6 and 24 hours, although no combinatorial effect of IL-1β and IFN-γ on TLR2 mRNA was observed. However, combined stimulation of A549 cells with IL-1β+IFN-γ+Dex profoundly induced TLR2 mRNA to 19- and 69-fold at 6 and 24 hours, respectively (P ≤ 0.001 and 0.01, respectively) (Fig. 2A, upper panels). Such induction of TLR2 expression was greater (all P ≤ 0.001) than was evident for IL-1β, IFN-γ, or dexamethasone alone at both 6 and 24 hours. Moreover, comparison of the sum of the effects of single treatments with that of the effect of the combined stimulus showed marked (P ≤ 0.05 and 0.01, respectively) supra-additivity between the effects of IL-1β, IFN-γ, and dexamethasone at 6 and 24 hours (Fig. 2A, lower panels). Furthermore, the addition of IL-1β, dexamethasone, or IFN-γ to each of the three dual treatments led to reproducible and statistically significant increases in TLR2 mRNA expression at both 6 and 24 hours (Fig. 2A; Supplemental Fig. 1B). Thus, clear positive combinatorial effects between IL-1β, IFN-γ, and dexamethasone on TLR2 mRNA expression were revealed in A549 cells.
Effect of IL-1β, IFN-γ and dexamethasone on TLR2 mRNA in human pulmonary epithelial cells. (A) A549 cells or (B) pHBECs grown in submersion culture were either not treated or treated with IL-1β (1 ng/ml), IFN-γ (10 ng/ml), and/or Dex (1 µM) as indicated. Cells were harvested at 6 or 24 hours for quantitative polymerase chain reaction analysis of TLR2 and GAPDH. (Upper graphs) TLR2 data from N independent experiments were normalized to GAPDH, expressed as fold of untreated, and mean ± S.D. were plotted as bar graphs with overlaid scatter plots. Significance was tested using ANOVA with a Tukey post hoc test. Asterisks (*, **, or ***) represent significance relative to untreated or as indicated. Hash (#, ##, or ###) represent significance relative to IL-1β+IFN-γ+Dex. (Lower graphs) The sum of each effect (i.e., fold − 1) within a group of treatments (sum) and the effect (fold − 1) for each combination treatment (comb) is expressed as mean ± S.D. and plotted as bar graphs overlaid with scatter plots. Significance was tested using paired t test. *, #P ≤ 0.05; **, ##P ≤ 0.01; ***, ###P ≤ 0.001.
Similarly, the effects of IL-1β, IFN-γ, and glucocorticoid on TLR2 mRNA were investigated in pHBECs. In submersion cultures of pHBECs, TLR2 mRNA was modestly induced by IL-1β, IFN-γ, and dexamethasone alone at both 6 and 24 hours (Fig. 2B, upper panels). Treatment with IL-1β+Dex induced (P ≤ 0.01) TLR2 mRNA at 6 hours, and IFN-γ+Dex induced (P ≤ 0.001) TLR2 mRNA at 24 hours (Fig. 2B, upper panels). Although combined stimulation with IL-1β+IFN-γ showed little increase in TLR2 mRNA at 6 and 24 hours, dexamethasone enhanced (P ≤ 0.001) the IL-1β+IFN-γ–induced TLR2 mRNA at both times (Fig. 2B, upper panels). Moreover, IL-1β+IFN-γ+Dex–induced TLR2 mRNA to 63- and 56-fold at 6 and 24 hours, respectively, and this was more (both P ≤ 0.01) than each of IL-1β, IFN-γ, or dexamethasone alone. Comparison of the effects of the combination treatments (comb), IL-1β+Dex, IFN-γ+Dex, or IL-1β+IFN-γ+Dex, to the sum of the effects of the respective individual treatments (sum) revealed obvious (all P ≤ 0.05) supra-additivity at both 6 and 24 hours (Fig. 2B. lower panels).
In ALI cultures of pHBECs, TLR2 mRNA was modestly induced by IL-1β and budesonide alone, and clear combinatorial effects, with supra-additivity (P ≤ 0.05), between IL-1β and budesonide were observed at both 6 and 24 hours (Supplemental Fig. 2B). Although IFN-γ barely induced TLR2 expression in the ALIs, cotreatment with IL-1β led to increased (P ≤ 0.05) TLR2 mRNA at both 6 and 24 hours (Supplemental Fig. 2B, upper panels). Similarly, IFN-γ+Bud cotreatment increased (P ≤ 0.001) TLR2 mRNA above baseline. Moreover, budesonide enhanced (P ≤ 0.01) IL-1β+IFN-γ–induced TLR2 mRNA to 11.6- and 7.8-fold at 6 and 24 hours, respectively. Thus, at both times, TLR2 mRNA induced by the combined stimulation with IL-1β+IFN-γ+Bud was higher (all P ≤ 0.01) than by IL-1β, IFN-γ, or budesonide alone (Supplemental Fig. 2B, upper panels). Indeed, comparison of the sum of the effects of IL-1β, IFN-γ, and budesonide alone (sum) with the effect of their combined stimulation (comb) revealed supra-additivity between IL-1β+IFN-γ+Bud at 24 hours (P ≤ 0.05) but not at 6 hours (Supplemental Fig. 2B, lower panels). However, this synergistic effect was no more than for IL-1β+Bud combination, i.e., IFN-γ had no additional effect over IL-1β+Bud combination.
As GAPDH expression may vary with some treatments, cycle threshold (CT) values to show GAPDH expression for all treatments are shown (Supplemental Fig. 3, A–C). In each case, this was unaffected by IL-1β, IFN-γ, or dexamethasone treatments in A549, pHBECs, and ALI cultures.
IKK Involvement in IL-1β+IFN-γ+Dex–Induced TLR2 Expression
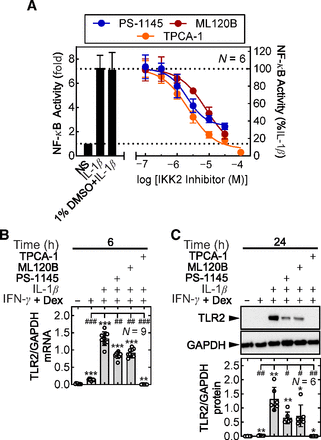
Prior work showed a role for NF-κB in the induction of TLR2 by IL-1β plus glucocorticoid (Bansal et al., 2022). Here, a role for the canonical IKK pathway, involving IKK2, in the IL-1β+IFN-γ+Dex–induced TLR2 expression was tested in A549 cells using the IKK2-selective inhibitors PS-1145, ML120B, and TPCA-1 (Gamble et al., 2012). For each compound, concentrations of 30 µM produced maximal inhibition of IL-1β–induced NF-κB luciferase reporter activity (Fig. 3A; Supplemental Fig. 4A).
Effect of IKK inhibitors on the enhancement of IFN-γ+Dex–induced TLR2 expression by IL-1β. (A) A549 cells containing the NF-κB reporter 6κBtk.luc were either not treated or pretreated with 1% DMSO or indicated concentrations of PS-1145, ML120B, or TPCA-1 for 90 minutes prior to stimulation with IL-1β (1 ng/ml). Cells were harvested for luciferase assay. Data from N independent experiments was expressed as fold (bars) and percent IL-1β (lines) and plotted as mean ± S.D. Scatterplots showing individual data points appear as Supplemental Fig. 4A. (B) A549 cells were pretreated with PS-1145, ML120B, or TPCA-1, each at 30 µM, for 1.5 hours prior to stimulation with IFN-γ (10 ng/ml) plus Dex (1 µM) and IL-1β (1 ng/ml) as indicated. Cells were harvested at 6 hours for RNA extraction and quantitative polymerase chain reaction analysis of TLR2 and GAPDH mRNA. (C) Cells were treated as in (B) and harvested at 24 hours for western blot analysis of TLR2 and GAPDH protein. (B and C) TLR2 data, from N independent experiments, were normalized to GAPDH and are expressed as mean ± S.D. and plotted as bar graphs overlaid with scatter plots. Significance was assessed by ANOVA with a Tukey multiple comparison test. Differences from untreated: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; from IL-1β+IFN-γ+Dex: #P ≤ 0.05; ##P ≤ 0.01; ###P ≤ 0.001.
TLR2 mRNA and protein expression were modestly induced by IFN-γ+Dex, at 6 and 24 hours, and these effects were significantly enhanced (P ≤ 0.001 and 0.01, respectively) by IL-1β (Fig. 3, B and C). In cells pretreated with 30 μM PS-1145, ML120B, or TPCA-1, the enhancement of IFN-γ+Dex–induced TLR2 mRNA and protein by IL-1β were reduced (all P ≤ 0.05) by each compound (Fig. 3, B and C). In each case, PS-1145 and ML120B produced partial inhibition, whereas TPCA-1 resulted in more complete inhibition of TLR2 mRNA and protein expression. Neither of these compounds affected the cell viability in the experimental conditions employed (Supplemental Fig. 4B). These data support a role, albeit possibly only partial, for IKK2 in the induction of TLR2 expression by IL-1β+IFN-γ+Dex.
Effects of IFN-γ on the possible activation of NF-κB were also examined. Treatment of A549 cells with IFN-γ did not elicit serine 32 and 36 phosphorylation of IκBα (Supplemental Fig. 4C). Likewise, IFN-γ alone did not induce NF-κB reporter activity and showed no effect on IL-1β–induced NF-κB reporter activity whether in the absence or presence of dexamethasone (Supplemental Fig. 4D, top panel). Thus, although IKK2 is implicated in the induction of TLR2 expression by IL-1β+IFN-γ+Dex, there was no effect of IFN-γ on activation of the NF-κB pathway or the induction of NF-κB transcriptional activity.
Involvement of GR in IL-1β+IFN-γ+Dex–Induced TLR2 Expression
A role for GR in the synergistic induction of TLR2 expression by IL-1β plus glucocorticoid was previously documented (Bansal et al., 2022). In the current study, the role of GR in the induction of TLR2 expression by IL-1β+IFN-γ+Dex was tested using the competitive GR antagonist Org34517 (Peeters et al., 2004; Joshi et al., 2015a). TLR2 mRNA and protein were both modestly induced by IL-1β+IFN-γ at 6 and 24 hours, respectively (Fig. 4, A and B; Supplemental Fig. 5, A and B). Increasing the concentration of dexamethasone markedly enhanced IL-1β+IFN-γ–induced TLR2 mRNA at 6 hours and protein at 24 hours. In each case, these enhancements by dexamethasone were competitive with Org34517, at 1 μM, as shown by rightward shifts in the response curve to dexamethasone (Fig. 4, A and B). Schild analysis to determine the negative logarithm of the molar concentration of antagonist that causes a twofold shift of the concentration-response curve for agonist, or pA2 value, resulted in pA2 values of 8.07 ± 0.16 and 7.85 ± 0.29 for TLR2 mRNA and protein, respectively, which is consistent with prior reports of competitive antagonism at GR. IL-1β+IFN-γ–induced TLR2 mRNA and protein were unaffected by Org34517.
Effect of Org34517 (Org) on the enhancement of IL-1β+IFN-γ–induced TLR2 expression by dexamethasone. A549 cells were pretreated with Org34517 (1 µM) for 1 hour prior to stimulation with IL-1β (1 ng/ml) plus IFN-γ (10 ng/ml) in the absence or presence of various concentrations of Dex. (A) After 6 hours, cells were harvested for RNA extraction and quantitative polymerase chain reaction analysis of TLR2 and GAPDH mRNA. (B) At 24 hours, cells were harvested for western blot analysis of TLR2 and GAPDH. In each case, TLR2 data, from N independent experiments, were normalized to GAPDH and are plotted as means ± S.D. Significance between treatments without and with Org was assessed by paired t test (two tailed). Note: unpaired t test was used at 0.1 µM Dex due to one sample failure in the western blotting. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. For illustration, curves were fitted following Schild analysis that assumed a common maximal effect (Emax) and a Schild slope factor of 1, with the lower asymptote fixed as the response to IL-1β+IFN-γ. This produced pA2 values of 8.07 (± 0.16) and 7.85 (± 0.29) for TLR2 mRNA and protein, respectively. Scatterplots for all data in (A) and (B) are shown as Supplemental Fig. 5, A and B.
Possible effects of IL-1β and IFN-γ on GRE-dependent transcriptional activity were investigated using A549 cells harboring a 2×GRE.luc reporter. GRE reporter activity was not induced by either IL-1β or IFN-γ and following induction by dexamethasone at 6 hours; reporter activity also remained largely unaffected by IL-1β and IFN-γ treatments (Supplemental Fig. 4D, middle panel). Thus, although the antagonism by Org34517 supports a role for GR in the induction of TLR2 by IL-1β+IFN-γ+Dex, GRE reporter activity was unaffected by IL-1β and/or IFN-γ, suggesting that an enhancement of simple GRE-dependent transcription does not directly contribute to the observed synergy in respect of TLR2 expression.
Activation of JAK-STAT Signaling by IFN-γ
STAT1 is the major transcription factor involved in IFN-γ–dependent transcriptional responses that occur via conventional JAK-STAT signaling (Schroder et al., 2004). To explore possible activation of STAT1, IFN-γ–induced phosphorylation of STAT1 at Y701 and S727, which are suggested to be involved in nuclear accumulation and transcriptional activation, respectively, were examined (Sadzak et al., 2008). Treatment of A549 cells with IFN-γ at 10 ng/ml induced rapid phosphorylation of STAT1 at Y701, with statistical significance observed from 15 minutes onwards (Fig. 5A; Supplemental Fig. 6A). This peaked at 1 hour before declining slightly by 6 and 24 hours. Phosphorylation of STAT1 at S727 was detectible in unstimulated cells, and this was increased (P ≤ 0.05) by 1 hour post IFN-γ treatment (Fig. 5B; Supplemental Fig. 6B). This phosphorylation declined slightly by 6 hours but was further elevated at 24 hours. However, although total STAT1 levels were unaffected by IFN-γ treatment at times until 6 hours, there was a ∼twofold (P ≤ 0.05) increase in total STAT1 at 24 hours (Fig. 5B; Supplemental Fig. 6B). This effect largely accounts for the increase in S727 phosphorylation observed at 24 hours. Overall, these data indicate activation of STAT1 by IFN-γ.
Effect of IL-1β, glucocorticoid, and IFN-γ on STAT1 phosphorylation. A549 cells were either untreated or treated with IFN-γ (10 ng/ml) prior to harvesting for western blot analysis at the indicated times. Representative blots of (A) phospho-STAT1 (P-STAT1 Y701) and GAPDH and (B) P-STAT1 (S727), total STAT1, and GAPDH from N independent experiments are shown. In each case, densitometric data normalized to GAPDH are plotted as means ± S.D. (error bars). Significance relative to untreated at 15 minutes was assessed by ANOVA with a Dunnett’s multiple comparison test. (C) A549 cells were either not treated or treated with IL-1β (1 ng/ml), IFN-γ (10 ng/ml), and/or Dex (1 µM) as indicated. Cells were harvested at 1 hour for western blot analysis of P-STAT1 (Y701 and S727) and GAPDH. Representative blots of P-STAT1 (Y701 & S727) and GAPDH from N independent experiments are shown. Densitometric data for P-STAT1 (Y701) (upper graph) and P-STAT1 (S727) (lower graph) were normalized to GAPDH, expressed as means ± S.D., and plotted as bar graphs overlaid with scatter plots. Significance relative to untreated (*) or IL-1β+IFN-γ (#) was tested using ANOVA with a Tukey post hoc test. *, #P ≤ 0.05; **, ##P ≤ 0.01; ***, ###P ≤ 0.001. Scatterplots for all data in (A) and (B) are shown as Supplemental Fig. 6.
The effects of IL-1β, IFN-γ, and dexamethasone were tested on phosphorylation of STAT1 at Y701 and S727 at 1 hour. STAT1 phosphorylation at Y701 was not induced by IL-1β, either in the presence or absence of dexamethasone (Fig. 5C, upper panels). IFN-γ strongly (P ≤ 0.05) induced STAT1 phosphorylation at Y701, and this was unaffected by the addition of IL-1β and/or dexamethasone (Fig. 5C, upper panels). Although dexamethasone did not induce STAT1 S727 phosphorylation, IL-1β alone induced (P ≤ 0.01) levels of S727 phosphorylated STAT1 that remained high with IL-1β+Dex cotreatment (Fig. 5C, lower panels). IFN-γ alone also induced (P ≤ 0.01) STAT1 S727 phosphorylation, and this was unchanged with IFN-γ+Dex cotreatment (Fig. 5C, lower panels). Combination of IL-1β+IFN-γ increased S727 STAT1 phosphorylation to a higher level than either IL-1β or IFN-γ alone, both P ≤ 0.01, and this was unaffected by dexamethasone (Fig. 5C, lower panels). Total STAT1 expression was not affected by IL-1β, IFN-γ, or dexamethasone whether alone or in combination (Supplemental Fig. 7A). Overall, these data show increased STAT1 phosphorylation at Y701 and S727 following IFN-γ treatment. S727 phosphorylation was also increased by IL-1β, and this appeared to be additive with the effect of IFN-γ. In each case, dexamethasone, alone or in combination with IL-1β and/or IFN-γ, did not affect STAT1 phosphorylation at Y701 and S727.
To test for possible JAK involvement, the JAK inhibitors AZD1480, barcitinib, and ruxolitinib were tested on the ability of IFN-γ to induce STAT1 phosphorylation. IFN-γ–induced phosphorylation of STAT1 at Y701 and S727 was prevented by each JAK inhibitor in a concentration-dependent manner (Fig. 6, A–C; Table 1). Inserts in Fig. 6C provide the log EC50 values obtained for each pair of curves describing the effect of each compound on S727 and Y701 phosphorylated STAT1 in each individual experiment (Fig. 6C). Both AZD1480 and baricitinib revealed greater (P ≤ 0.01 and 0.05, respectively) sensitivity of inhibition in respect of S727 compared with Y701 phosphorylation of STAT1. With ruxolitinib, this effect was much reduced, and although reaching P = 0.049, it should be noted that one set of data were excluded (Supplemental Fig. 8). The apparent difference in log EC50 values should therefore be treated with caution. Total STAT1 expression remained unaffected by the various concentrations of JAK inhibitors in the presence of IFN-γ (data not shown). Furthermore, unstimulated A549 cells showed lower levels of STAT1 phosphorylation at S727, and these were unaffected by 1 μM of each of the JAK inhibitors (Fig. 6D, left panel). IL-1β treatment increased STAT1 S727 phosphorylation compared with baseline, which also remained unaffected by the JAK inhibitors (Fig. 6D, left panel). Treatment with IFN-γ increased STAT1 phosphorylation at S727, and this was completely abrogated to baseline by the JAK inhibitors (Fig. 6D, right panel). Treatment with IL-1β+IFN-γ induced S727 phosphorylation to a greater level than IFN-γ alone, and each JAK inhibitor partially, but significantly (all P ≤ 0.05), reduced this response (Fig. 6D, right panel). As stated above, STAT1 phosphorylation at Y701 was not observed in unstimulated or IL-1β–treated A549 cells, and this was not changed by the JAK inhibitors (data not shown). IFN-γ and IL-1β+IFN-γ treatments strongly induced similar levels of STAT1 Y701 phosphorylation, and this induction was completely prevented by all three JAK inhibitors (Fig. 6E). Total STAT1 expression was unaffected by IL-1β, IFN-γ, or IL-1β+IFN-γ treatments either in the presence or absence of JAK inhibitors (Supplemental Fig. 7B). Although these data support the involvement of JAKs in the activation of STAT1 by IFN-γ, whether in the presence or absence of IL-1β, the apparent differences in selectivity on phosphorylation of STAT1 at Y701 versus S727 raise the possibility of mechanistic differences in each process.
Effect of JAK inhibitors on STAT1 phosphorylation. (A) A549 cells were pretreated with AZD1480, barcitinib and ruxolitinib at the indicated concentrations for 30 minutes, followed by stimulation with IFN-γ (10 ng/ml) for 1 hour. Cells were harvested for western blot analysis of phospho-STAT1 (P-STAT1) and GAPDH. Representative blots for P-STAT1 (Y701), P-STAT1 (S727), and GAPDH from five independent experiments are shown. (B) Densitometric data for Y701 P-STAT1 and S727 P-STAT1 from western blots shown in (A) were normalized to GAPDH and plotted as mean ± S.D. (C) Data were expressed as percent IFN-γ effect for each of AZD1480, barcitinib, and ruxolitinib and plotted as means ± S.D. Log EC50 of each compound for the inhibition of P-STAT1 (Y701) and P-STAT1 (S727) is expressed as mean ± S.D. and shown as scatter plots in the inserts within each graph of (C). Note: one dataset was excluded for ruxolitinib, and the full data are shown as Supplemental Fig. 8. (D and E) A549 cells were either not treated or pretreated with AZD1480, barcitinib, and ruxolitinib, each at 1 µM, for 1 hour prior to stimulation with IL-1β (1 ng/ml) and IFN-γ (10 ng/ml) as indicated. Cells were harvested at 1 hour for western blot analysis of P-STAT1 and GAPDH. Representative blots for P-STAT1 (Y701 & S727) and GAPDH from N independent experiments are shown. Densitometric data for (D) P-STAT (S727) and (E) P-STAT1 (Y701) were normalized to GAPDH, expressed as means ± S.D., and plotted as bar graphs overlaid with scatter plots. Significance relative to untreated (*), IFN-γ (#), or IL-1β + IFN-γ ($) was tested using ANOVA with a Tukey post hoc test. *, #, $P ≤ 0.05; **, ##, $$P ≤ 0.01; ***, ###, $$$P ≤ 0.001.
Concentration-response analysis of JAK inhibitors on IFN-γ–induced STAT1 phosphorylation and STAT1 reporter activity
pEC50 values of the three JAK inhibitors, AZD1480, barcitinib, and ruxolitinib, on inhibition of P-Y701 STAT1, P-S727 STAT1, and STAT1 reporter activity in IFN-γ–treated A549 cells are shown (related to Figs. 6, A–C and 7E). Data from four to five independent experiments are shown.
Activation of STAT1-Driven Transcription by IFN-γ
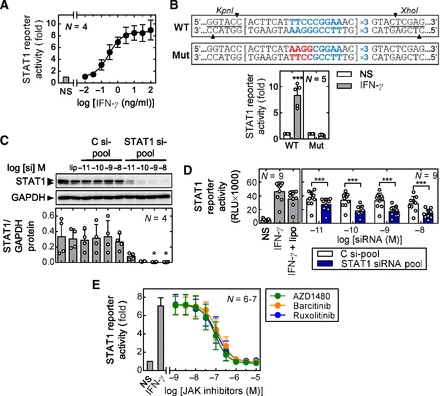
To explore the transcriptional activation of STAT1 by IFN-γ, A549 cells harboring a luciferase reporter containing three consensus STAT1 motifs, corresponding to a conventional GAS, upstream of a luciferase gene driven by a basal promoter (3×STAT1.luc), were stimulated with various concentrations of IFN-γ (Fig. 7A; Supplemental Fig. 9A). Reporter activity, measured at 6 hours, was induced by IFN-γ in a concentration-dependent manner (EC50 = 0.28 ng/ml), with maximal activity occurring at ∼10 ng/ml. This correlated with the concentration of IFN-γ that showed maximal synergy with IL-1β and glucocorticoid on the induction of TLR2 expression. To confirm a role for the consensus STAT1 motifs in the induction of reporter activity, A549 cells were stably transfected with a reporter construct in which the 3×STAT1 motifs were replaced with mutated sites (Fig. 7B, top panel). As above, IFN-γ, at 10 ng/ml, induced (P ≤ 0.001) 3×STAT1.luc reporter activity, whereas cells containing the mutated STAT1 reporter showed no enhancement of reporter activity (Fig. 7B, bottom panel). These data indicate that the consensus STAT1 motif conferred IFN-γ inducibility. To confirm a role for STAT1 in IFN-γ–induced reporter activity, A549 cells were transfected with various concentrations of four different STAT1-targeting siRNAs, or a pool of STAT1 siRNAs, alongside a pool of nontargeting siRNAs as a control. Although the nontargeting control siRNAs had no effect, each individual STAT1-targeting siRNA dramatically reduced STAT1 protein expression, with maximal effects apparent at 1 and 10 nM (Fig. 7C; Supplemental Fig. 10). A pool of these four STAT1-targeting siRNAs also reduced (P ≤ 0.05) STAT1 protein expression by ∼90% at both 1 and 10 nM (Fig. 7C). Furthermore, IFN-γ–induced STAT1 reporter was unaffected by various concentrations of the control siRNA pool, whereas the STAT1 siRNA pool markedly diminished (P ≤ 0.001) reporter activity at all siRNA concentrations (Fig. 7D). This confirms the involvement of STAT1 in the activation of this reporter by IFN-γ. Moreover, in A549.3×STAT1.luc cells pretreated with various concentrations of the JAK inhibitors AZD1480, barcitinib, and ruxolitinib, IFN-γ–induced STAT1 transcriptional activity was abrogated in a concentration-dependent manner (pEC50 = 7.05 ± 0.22, 6.86 ± 0.21, and 6.98 ± 0.19 for AZD1480, barcitinib, and ruxolitinib, respectively) (Fig. 7E; Supplemental Fig. 9B). These data confirm that IFN-γ activates a JAK pathway and leads to STAT1-dependent transcription via consensus STAT1 motifs.
Characterization of a STAT1-driven luciferase reporter. (A) A549 cells stably transfected with a luciferase reporter containing three copies of a consensus STAT1-binding motif [shown in (B)] were either not stimulated (NS) or stimulated with the indicated concentrations of IFN-γ prior to harvesting for luciferase assay after 6 hours. Luciferase activity from N independent experiments was expressed as fold of NS and plotted as mean ± S.D. (B) (Upper panel) reporter construct showing three copies of the consensus/wild-type (WT; bold blue) or mutant (Mut; bold blue + red) STAT1-binding motif is shown. KpnI and XhoI restriction sites used to clone the construct in pGL3.TATA.neo vector are underlined, with cut sites shown as black arrowheads. (Lower panel) A549 cells were stably transfected with the WT or Mut STAT1 reporter constructs and either NS or treated with IFN-γ (10 ng/ml) for 6 hours followed by harvesting for luciferase assay. Luciferase activity from N independent experiments was expressed as fold of NS, and mean ± S.D. was plotted as bar graphs overlaid with scatter plots. Significance relative to untreated cells was determined using paired t test. (C) A549 cells were either NS or incubated with lipid control (lip) alone or with the indicated concentrations of either control siRNA pool (C si-pool), or STAT1 siRNA pool (STAT1 si-pool) for 48 hours. Cells were then incubated overnight with serum-free media prior to harvesting for western blot analysis. Representative blots of STAT1 and GAPDH from N independent experiments are shown. Data obtained from densitometry analysis were normalized to GAPDH, and mean ± S.D. was plotted as bar graphs overlaid with scatter plots. Significance relative to C si-pool at each siRNA concentration was tested by paired t test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (D) A549 cells stably transfected with the WT STAT1 reporter construct were treated with lipid or siRNAs as described in (C) prior to stimulation with IFN-γ (10 ng/ml) for 6 hours. Cells were harvested for luciferase assay, and raw luciferase units (RLU) from N independent experiments were expressed as mean ± S.D. and plotted as bar graphs overlaid with scatter plots. Significance relative to C si-pool at each siRNA concentration was tested by paired t test. (E) A549 cells stably transfected with the WT STAT1 reporter construct were either NS or pretreated with the indicated concentrations of AZD1480, barcitinib, or ruxolitinib for 30 minutes followed by stimulation with IFN-γ (10 ng/ml). Cells were harvested for luciferase assay after 6 hours. Luciferase activity from N independent experiments was expressed as fold of NS and plotted as mean ± S.D. Scatterplots for all data in (A) and € are shown as Supplemental Fig. 9.
Involvement of JAK Signaling in IL-1β+IFN-γ+Dex–Induced TLR2 Expression
To investigate a possible role for JAKs in IL-1β+IFN-γ+Dex–induced TLR2 expression, A549 cells were pretreated for 1 hour with AZD14580, barcitinib, and ruxolitinib (each at 1 μM) prior to stimulation with IL-1β, IFN-γ, and dexamethasone. TLR2 mRNA and protein were modestly induced by IL-1β+Dex at 6 and 24 hours, respectively, and this was markedly enhanced (both P ≤ 0.01) by IFN-γ (Fig. 8). AZD14580, barcitinib, and ruxolitinib each reduced the enhancement of IL-1β+Dex–induced TLR2 mRNA by IFN-γ back to the level achieved by IL-1β+Dex (Fig. 8A). As similar data were also obtained for TLR2 protein (Fig. 8B), these data support involvement of JAK signaling in the enhancement of IL-1β+Dex–induced TLR2 expression by IFN-γ.
Effect of JAK inhibitors on the enhancement of IL-1β+Dex–induced TLR2 expression by IFN-γ. A549 cells were either not stimulated or pretreated with AZD1480, barcitinib, or ruxolitinib, each at 1 µM, for 1 hour prior to stimulation with IL-1β (1 ng/ml) plus Dex (1 µM) and IFN-γ (10 ng/ml) as indicated. (A) Cells were harvested at 6 hours for RNA extraction and quantitative polymerase chain reaction analysis of TLR2 and GAPDH mRNA. (B) Cells were harvested at 24 hours for western blot analysis of TLR2 and GAPDH protein. In each case, TLR2 data, from N independent experiments, were normalized to GAPDH, and mean ± S.D. were plotted as bar graphs overlaid with scatter plots. Significance was assessed by ANOVA with a Tukey multiple comparison test. Differences from untreated: **P ≤ 0.01; ***P ≤ 0.001; from IL-1β+IFN-γ+Dex: ##P ≤ 0.01; ###P ≤ 0.001.
The effect of IL-1β and dexamethasone on IFN-γ–induced STAT1 transcriptional activity was also investigated using the A549.3×STAT1.luc cells. IL-1β and dexamethasone did not induce STAT1 reporter activity. IFN-γ treatment induced strong activation of STAT1 reporter that was modestly, but significantly (P ≤ 0.05), repressed by IL-1β and largely unaffected by dexamethasone (Supplemental Fig. 4D, lower panel). Since IFN-γ–induced STAT1 reporter activity was either not affected or reduced by IL-1β and/or dexamethasone, simple effects on STAT1-dependent transcription by combined stimulation of IL-1β+IFN-γ+Dex is unlikely to explain the synergy observed between IL-1β, IFN-γ, and dexamethasone at inducing TLR2 expression.
Roles of p65, GR, and STAT1 on TLR2 Expression Induced by IL-1β, IFN-γ, and Dexamethasone
As shown in Supplemental Fig. 4D, IL-1β–induced NF-κB reporter activity was unaltered by IFN-γ and/or dexamethasone. Similarly, as seen above, dexamethasone-induced GRE reporter activity was not affected by IL-1β and/or IFN-γ. Likewise, IFN-γ–induced STAT1 reporter activity was also unaffected by IL-1β and/or dexamethasone. Therefore, the effects of IL-1β on GR and STAT1 signaling, or of dexamethasone on NF-κB and JAK-STAT pathway, or of IFN-γ on the NF-κB or GR signaling, do not explain the synergy between IL-1β, IFN-γ, and dexamethasone at the level of TLR2 expression. Nevertheless, the involvement of p65, GR, and STAT1 transcription factors, as the primary factors activated by each pathway, seems highly plausible in the synergy in TLR2 expression by combined stimulation with IL-1β, IFN-γ, and dexamethasone. This was tested by knocking down the expression of each factor. To confirm the knockdown of p65, A549 cells were transfected with various concentrations of four different p65-targeting siRNAs, or a pool of p65 siRNAs, alongside a pool of nontargeting siRNAs as a control. Although the nontargeting control siRNAs had no effect on p65 protein expression, each individual p65-targeting siRNA, as well the p65 siRNA pool, dramatically reduced p65 protein expression at 1 and 10 nM (Supplemental Fig. 11A). NF-κB reporter activity was also tested to assess the effectiveness of the p65 knockdown. Thus, increasing concentrations of each of the p65 siRNAs, along with the p65 siRNA pool, reduced the IL-1β–induced NF-κB reporter activity, where the greatest effect was observed at 1 and 10 nM, and the nontargeting control siRNAs had no effect (Supplemental Fig. 11B). To achieve knockdown of GR, siRNAs used in a prior study were employed (Thorne et al., 2023). Further, the role of each of p65, GR, and STAT1 was tested on TLR2 expression. Therefore, A549 cells were transfected with siRNA pools targeting each of p65, GR, and/or STAT1, followed by treatment with various combinations of IL-1β, IFN-γ, and dexamethasone for 24 hours. With IL-1β+Dex, the control and STAT1-targeting siRNAs had no marked effect on TLR2 protein, whereas marked reductions (P ≤ 0.01 and 0.05, respectively) in TLR2 protein were observed following knockdown of either p65 and/or GR (Fig. 9, A and B). This confirms roles for both p65 and GR in the induction of TLR2 expression by IL-1β+Dex. Likewise, in cells treated with IFN-γ+Dex, the knockdown of STAT1 and GR reduced (both P ≤ 0.01) TLR2 protein levels, whereas control and p65 siRNA pools had no effect (Fig. 9, A and C). Therefore, STAT1 and GR appear to be involved in IFN-γ+Dex–induced TLR2 expression. Similarly, in cells treated with the combination of IL-1β+IFN-γ+Dex, whereas nontargeting control siRNAs had no significant effect, the p65 and GR siRNA pools almost completely abolished (both P ≤ 0.001) the IL-1β+IFN-γ+Dex–induced TLR2 protein (Fig. 9D). The STAT1 siRNA pool also significantly (P ≤ 0.05), but only partially, reduced TLR2 expression induced by IL-1β+IFN-γ+Dex (Fig. 9 D). These data confirm roles for p65, GR, and STAT1 in the induction of TLR2 expression by IL-1β, IFN-γ, and dexamethasone in combination. Finally, in the case of IL-1β+IFN-γ–treated A549 cells, only the p65 siRNA pool reduced (P ≤ 0.05) TLR2 protein, whereas the controls, GR, and STAT1-targeting siRNAs had no effect on TLR2 expression (Fig. 9, A and E). This suggests that p65, but not STAT1, appears to be involved in the TLR2 expression induced by IL-1β+IFN-γ.
Effect of p65, GR, and STAT1 knockdown on IL-1β, IFN-γ, and dexamethasone-induced TLR2 expression in A549 cells. (A) A549 cells were incubated with pools of either control siRNAs (C si-pool), p65 siRNAs (p65 si-pool), GR siRNAs (GR si-pool), or STAT1 siRNAs (STAT1 si-pool) (1 nM each) for 48 hours. Cells were then incubated overnight with serum-free media prior to harvesting for western blot analysis. Representative blots of p65, GR, STAT1, and GAPDH from five independent experiments are shown. (B–E) A549 cells were incubated with siRNA pools as in (A), followed by treatment with IL-1β (1 ng/ml), IFN-γ (10 ng/ml), and/or Dex (1 µM), as indicated, for 24 hours. Cells were harvested for western blot analysis. Representative blots of TLR2 and GAPDH from N independent experiments are shown. Densitometric data were normalized to GAPDH, expressed as fold of untreated, and mean ± S.D. were plotted as bar graphs overlaid with scatter plots. Significance relative to C si-pool in each case was tested using ANOVA with a Dunnett’s post hoc test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Discussion
Glucocorticoids have complex impacts on inflammation and variously downregulate or enhance gene expression (Busillo and Cidlowski, 2013; Cruz-Topete and Cidlowski, 2015; Amrani et al., 2020). Aside from metabolic effects, many glucocorticoid-enhanced genes, by controlling signaling and gene expression, are regulatory, whereas others are proinflammatory (Busillo et al., 2011; Newton et al., 2017). Mechanisms and relevance for these different effects remain poorly understood but are required to build a more comprehensive understanding of glucocorticoid action that considers both anti-inflammatory effects with potential enhancements of inflammatory responses. The current study focused on synergy between glucocorticoid and cytokines to induce TLR2 expression in human pulmonary epithelial cells. Consistent with prior reports, dexamethasone when combined with each of IL-1β or IFN-γ supra-additively enhanced TLR2 expression in A549 cells (Shuto et al., 2002; Hermoso et al., 2004; Homma et al., 2004; Sakai et al., 2004; Hoppstädter et al., 2019; Bansal et al., 2022). Building from this, we report that in A549 cells and pHBECs, a clear three-way supra-additivity occurred between the glucocorticoid, IL-1β, and IFN-γ as a triple combination. At the mRNA level, the effect of this triple combination surpassed the responses to the individual and dual treatments. Indeed, occurring at maximally effective concentrations of each stimulus, this represents synergy between the three pathways that was readily apparent and replicated at both the mRNA and protein levels of TLR2 expression. Since all the current analyses were at 24 hours or less, further synergy could be observed at longer times, but this would require additional testing. Furthermore, replication of these effects in pHBECs grown in submersion culture supports A549 cells as a model for mechanistic investigations. Nevertheless, although pHBECs grown in ALI culture showed supra-additivity between IL-1β and glucocorticoid on TLR2 mRNA, effects of IFN-γ were not apparent. This discrepancy could be due to several reasons. For example, IL-1β plus glucocorticoid may induce maximal TLR2 expression such that combinatorial effects with IFN-γ require lower levels of IL-1β plus glucocorticoid stimulation. Moreover, signaling and the accessibility of regulatory elements responsible for TLR2 expression induced by IL-1β, IFN-γ, and glucocorticoid may differ between undifferentiated pHBECs in submersion culture and highly differentiated ALI cultures. Equally, disparities in the expression of necessary factors between undifferentiated and differentiated pHBECs could differentially regulate TLR2, whereas enhancer usage can certainly change with differentiation state (Natoli, 2010). In addition, differential contributions by the various differentiated ALI cell types may confound the overall effect. Although not explored, this differential responsiveness could be relevant to heighten host responses in damaged pseudostratified epithelia, where cells undergo dedifferentiation as a necessary part of healing and restoration of barrier function.
To better understand combinatorial effects between glucocorticoid, IL-1β, and IFN-γ, we identified key factors involved with each stimulus. The regulation of gene expression by IL-1β and IFN-γ commonly involves NF-κB and STAT1, respectively (Pahl, 1999; Schroder et al., 2004), and can produce profound transcriptional synergy (Hiroi and Ohmori, 2005). However, such synergy was not readily observed for TLR2. Rather, TLR2 synergies required the presence of glucocorticoid, prompting us to explore roles for NF-κB, STAT1, and GR. The two glucocorticoids, dexamethasone and budesonide, used in the current study are both GR selective and produce synonymous effects on gene expression (Hellal-Levy et al., 1999; Millan et al., 2011; Mostafa et al., 2019). By employing Org34517, a competitive GR antagonist, that, unlike RU496 (mifepristone) (Jewell et al., 1995; Chivers et al., 2004; Rider et al., 2018), may not lead to GR translocation (Peeters et al., 2008) and which shows a much-reduced level of partial agonism (Joshi et al., 2015a,b), we indicate a role for GR. Given possible off-target effects of antagonists on other nuclear hormone receptors, we also used silencing of GR to provide compelling evidence for GR involvement in the synergy between IL-1β, IFN-γ, and glucocorticoid. This aligns with glucocorticoid-induced GR binding to GRE-containing DNA regions upstream of human TLR2 to activate transcription (Bansal et al., 2022).
The NF-κB pathway was explored using maximally effective concentrations of the IKK2-selective inhibitors PS-1145, ML120B, and TPCA-1. Partial, but significant, reductions in IL-1β+IFN-γ+Dex–induced TLR2 expression by PS-1145 and ML120B, as well as the near complete inhibition by TPCA-1, support roles for the IKK2-NF-κB pathway. However, the partial effects of PS-1145 and ML120B, combined with their greater selectivity for IKK2 compared with TPCA-1 (Gamble et al., 2012), also raise the possible involvement of non-IKK2 pathways. Conversely, the greater inhibition of NF-κB–dependent transcription and TLR2 expression observed for TPCA-1 compared with PS-1145 and ML120B implies off-target effects for this compound. Indeed, reports that TPCA-1 prevents IFN-γ–induced responses, in the absence of NF-κB activation, support this concern (Tudhope et al., 2007). Nevertheless, following p65 silencing, the resultant loss of TLR2 expression when induced by IL-1β+IFN-γ+Dex, but not by IFN-γ+Dex, indicates a key role for NF-κB. Given p65 binding to the human TLR2 promoter (Bansal et al., 2022), the current data confirms p65/NF-κB in synergy between IL-1β, IFN-γ, and glucocorticoid.
IFN-γ treatment of A549 cells induced STAT1 phosphorylation at S727 and Y701, with Y701 phosphorylation markedly preceding that for S727. Although consistent with STAT1 activation (Wen et al., 1995; Sadzak et al., 2008), the delay in S727 phosphorylation compared with the rapid increase in Y701 phosphorylation suggests mechanistic differences. It is likely that IFN-γ receptor activation produces rapid JAK-dependent phosphorylation of Y701, facilitating STAT1 nuclear localization (Sadzak et al., 2008). Conversely, delayed S727 phosphorylation may involve an independent pathway or be consequent on prior Y701 phosphorylation. Notably, S727 phosphorylation was induced by IL-1β, and additive summation with the effect of IFN-γ suggests additional, likely non-JAK, pathways to phosphorylate S727. Indeed, mitogen activated protein kinases (MAPKs), which are activated by IL-1β in these cells (Shah et al., 2014), can phosphorylate S727 (Kovarik et al., 1999; Ramsauer et al., 2002; Zhang et al., 2004). However, in considering effects of IL-1β on S727 phosphorylation, it is salient that IFN-γ–induced STAT1/GAS reporter activity was reduced upon cotreatment with IL-1β and therefore questions a positive role for this phosphorylation event.
Although the JAK inhibitors AZD1480, barcitinib, and ruxolitinib were highly efficacious and similarly potent on IFN-γ–induced STAT1 reporter activity, their effects on STAT1 phosphorylation suggest site-specific differences. Specifically, S727 phosphorylation, compared with Y701 phosphorylation, appeared more sensitive to inhibition by AZD1480 and barcitinib relative to ruxolitinib. Various possibilities may explain this. Given the time lag between Y701 and S727 phosphorylation, it is possible that high levels of JAK activity are required for subsequent S727 phosphorylation. Therefore, modest inhibition of the relevant JAK could reduce S727 phosphorylation while having a reduced or no effect on Y701 phosphorylation. However, this hypothesis predicts parallel inhibitory effects of each inhibitor on each phosphorylation. This was likely not observed for ruxolitinib, and this raises the possibility that the differential selectivity of the inhibitors for JAK1, JAK2, JAK3, and Tyk2 could explain the data. Indeed, AZD1480 and baricitinib are relatively selective for JAK1 and JAK2, whereas ruxolitinib shows a less selective inhibition profile against the four JAKs (Fridman et al., 2010; Quintás-Cardama et al., 2010; Ioannidis et al., 2011). Therefore, different JAKs may plausibly produce these two phosphorylation events. This will need to be resolved by additional studies. Further, all three compounds revealed similar, but lower, potency on the STAT1 reporter. Thus, although STAT1 phosphorylation was relatively sensitive to JAK inhibition, the reporter system may show a “reserve” requiring higher levels of kinase inhibition to reduce reporter activity. This is consistent with the silencing of STAT1, where >90% loss of STAT1 protein produced just over 50% inhibition of reporter activity. Similar effects on other pathways involving kinase activation of transcriptional responses are observed, which, anecdotally, aligns with signal amplification and signaling reserves toward the distal parts of pathways.
Regarding synergies produced by IL-1β, IFN-γ, and glucocorticoid on TLR2 expression, we provide strong evidence for roles of pathways leading to NF-κB, STAT1, and GR. However, coactivation of these pathways had little effect on transcriptional activation produced by each factor acting via its respective consensus motif. This necessitates gene-specific explanations for the observed synergy. In prior studies, GR and p65 binding to upstream regions of the TLR2 locus was unaffected by cotreatments and suggested that the presence of both factors at the TLR2 promoter was sufficient for combinatorial effects (Bansal et al., 2022). However, these same promoter regions were unresponsive to IFN-γ (data not shown), indicating involvement of STAT1 at distinct genomic regions. Further studies are therefore required to delineate any such regions.
The current findings advance understanding of the mechanisms leading to synergy but do not address the functional significance of TLR2 synergy. Prior investigators have debated pro- and anti-inflammatory effects for TLR2 (Imasato et al., 2002; Homma et al., 2004; Winder et al., 2009; Hoppstädter et al., 2019). However, TLR2 agonism in cells showing IL-1β+Dex–induced TLR2 stimulated NF-κB reporter activity and induced CXCL8 and CCL5 expression (Bansal et al., 2022). This is consistent with other TLR and IL1 family receptors, which activate NF-κB, MAPKs, and inflammatory responses (O’Neill and Bowie, 2007). Likewise, in pHBECs treated with TNF or IFN-γ plus dexamethasone, TLR2 agonists induce expression of inflammatory genes that include IL6, CXCL8, and human β-defensin-2 (Hertz et al., 2003; Homma et al., 2004; Winder et al., 2009). Moreover, as TLR2 agonism reduced responsiveness to β2 agonists and glucocorticoids (Alkhouri et al., 2014; Rahman et al., 2016), its expression may be unhelpful in chronic inflammatory diseases. Indeed, TLR2 expression is widely documented as being elevated in severe asthma (Simpson et al., 2007; Singhania et al., 2018; Zhang et al., 2021), a condition in which patients are subjected to high-intensity therapies involving β2-adrenoceptor agonists and glucocorticoids. Conversely, enhanced TLR2 expression and activation of inflammatory responses may facilitate host defense responses, while glucocorticoids simultaneously suppress general inflammation and promote healing (Busillo and Cidlowski, 2013; Cruz-Topete and Cidlowski, 2015; Amrani et al., 2020). Thus, the cooperation between GR, NF-κB, and STAT1 to induce TLR2 expression may be important for normal host defense but could be undesirable in the (glucocorticoid) treatment of chronic inflammatory disease, including severe asthma, where the expression of cytokines such as IL-1β and IFN-γ is expected. Indeed, although priming of the innate immune response is reported for TLR2 agonists and may be enhanced in the presence of glucocorticoid (Alshaghdali et al., 2021; Deliyannis et al., 2021; Girkin et al., 2021), the antagonism of TLR2, or targeting of TLRs, has also been shown to reduce expression of inflammatory mediators (Su and Weindl, 2018; Thapa et al., 2023).
Acknowledgments
The authors would like to thank David Proud for his assistance with the provision of pHBECs. They also would like to acknowledge the Canadian Fund of Innovation (CFI) and the Alberta Science and Research Authority for providing an equipment and infrastructure grant to perform real-time PCR.
Data Availability
This article contains no datasets generated or analyzed during the current study.
Authorship Contributions
Participated in research design: Bansal, Newton.
Conducted experiments: Bansal, Kooi, Kalyanaraman, Gill, Thorne, Chandramohan, Necker-Brown, Mostafa, Milani.
Contributed new reagents or analytical tools: Leigh, Newton.
Performed data analysis: Bansal, Kooi, Kalyanaraman, Gill, Thorne, Chandramohan, Necker-Brown, Mostafa, Milani.
Wrote or contributed to the writing of the manuscript: Bansal, Leigh, Newton.
Footnotes
- Received June 18, 2023.
- Accepted September 29, 2023.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) [funding reference numbers: PJT 156310 (to R.N.) and 180480 (to R.N. and R.L.)] and a Natural Sciences and Engineering Research Council of Canada (NSERC) discovery [Grant RGPIN-2016-04549] (to R.N.); studentships (to A.B.): Eyes High Doctoral Scholarship and Eleanor Mackie Doctoral Scholarship in Women’s Health from University of Calgary. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data.
No author has an actual or perceived conflict of interest with the contents of this article.
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- ALI
- air-liquid interface
- AZD1480
- (S)-5-chloro-N2-(1-(5-fluoropyrimidin-2-yl)ethyl)-N4-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2, 4-diamine
- barcitinib
- 3-Azetidineacetonitrile, 1-(ethylsulfonyl)-3-[4-(7H-pyrrolo[2, 3-day]pyrimidin-4-yl)-1H-pyrazol-1-yl]-
- Bud
- budesonide
- Dex
- dexamethasone
- GAS
- γ-interferon–activated sequence
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element
- ICS
- inhaled corticosteroid
- IFN-γ
- interferon-γ
- IFN-γ+Dex
- IFN-γ plus dexamethasone
- IKK
- iκB kinase
- IL-1β
- interleukin-1β
- IL-1β+Bud
- IL-1β plus budesonide
- IL-1β+Dex
- IL-1β plus dexamethasone
- IL-1β+IFN-γ+Bud
- IL-1β plus IFN-γ plus budesonide
- IL-1β+IFN-γ+Dex
- IL-1β plus IFN-γ plus dexamethasone
- JAK
- Janus kinase
- MAPK
- mitogen activated protein kinase
- ML120B
- N-(6-Chloro-7-methoxy-9H-pyrido[3, 4-b]indol-8-yl)-2-methyl-3-pyridinecarboxamide dihydrochloride
- MTT
- 3-[4, 5-Dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide
- NF-κB
- nuclear factor κB
- Org34517
- Organon 34517
- pHBEC
- primary human bronchial epithelial cell
- PS-1145
- N-(6-chloro-9h-pyrido[3, 4-b]indol-8-yl)-3-pyridinecarboxamide
- ruxolitinib
- βR-cyclopentyl-4-(7H-pyrrolo[2, 3-day]pyrimidin-4-yl)-1H-pyrazole-1-propanenitrile
- siRNA
- short interfering RNA
- STAT
- signal transducer and activator of transcription
- TLR
- toll-like receptor
- TNF
- tumor necrosis factor
- TPCA-1
- 2-[(Aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide
- Copyright © 2023 by The Author(s)
This is an open access article distributed under the CC BY Attribution 4.0 International license.