Abstract
Etoposide is a widely used anticancer drug successfully used for the treatment of many types of cancer in children and adults. Its use, however, is associated with an increased risk of development of secondary acute myelogenous leukemia involving the mixed-lineage leukemia (MLL) gene (11q23) translocations. Previous studies demonstrated that the phenoxyl radical of etoposide can be produced by action of myeloperoxidase (MPO), an enzyme found in developing myeloid progenitor cells, the likely origin for myeloid leukemias. We hypothesized, therefore, that one-electron oxidation of etoposide by MPO to its phenoxyl radical is important for converting this anticancer drug to genotoxic and carcinogenic species in human CD34+ myeloid progenitor cells. In the present study, using electron paramagnetic resonance spectroscopy, we provide conclusive evidence for MPO-dependent formation of etoposide phenoxyl radicals in growth factor-mobilized CD34+ cells isolated from human umbilical cord blood and demonstrate that MPO-induced oxidation of etoposide is amplified in the presence of phenol. Formation of etoposide radicals resulted in the oxidation of endogenous thiols, thus providing evidence for etoposide-mediated MPO-catalyzed redox cycling that may play a role in enhanced etoposide genotoxicity. In separate studies, etoposide-induced DNA damage and MLL gene rearrangements were demonstrated to be dependent in part on MPO activity in CD34+ cells. Together, our results are consistent with the idea that MPO-dependent oxidation of etoposide in human hematopoietic CD34+ cells makes these cells especially prone to the induction of etoposide-related acute myeloid leukemia.
Introduction
Etoposide [VP-16, 4′-demethyl-epipodophyllotoxin-9-(4,6-O-ethylidene-β-d-gluco-pyranoside)] is a DNA topoisomerase II-targeting agent that has been used extensively as an anticancer agent to treat a variety of malignancies in adults and in children (Hande, 1998). However, the use of etoposide has been associated with an increased risk of developing secondary leukemias, especially acute myelogenous leukemia (t-AML), bearing translocations of the MLL gene at human chromosomal band 11q23 (Libura et al., 2005; Felix et al., 2006). Etoposide, which contains a hindered ring phenol, can be converted to phenoxyl radical forms by the action of peroxidases (Haim et al., 1987; Kagan et al., 1999). Because myeloid progenitor CD34+ cells in early stages of maturation contain the enzyme myeloperoxidase (MPO) (Strobl et al., 1993), we hypothesized that oxidative activation of the etoposide phenolic group by MPO may lead to MPO-catalyzed oxidative stress, including carcinogenic oxidative modification of DNA (Kagan et al., 2001). Hence, MPO expressed in CD34+ cells may make these myeloid progenitors especially sensitive to the leukemogenic action of etoposide.
MPO-induced oxidative stress is triggered by this enzyme's reactive intermediates, which have very high (1.35 V) oxidizing potential (Jantschko et al., 2005; Davies et al., 2008). In the presence of reducing substrates, particularly phenolic compounds such as etoposide, the one-electron oxidation catalyzed by MPO to yield phenoxyl radicals can in turn lead to interaction with a variety of cellular targets including lipids, thiols, ascorbate, proteins, and DNA (Zhang et al., 2002; Borisenko et al., 2004). Depending on the reactivity of the MPO-generated phenoxyl radicals, the oxidation of these cellular constituents may be directly or indirectly involved in MPO-driven oxidations and/or carcinogenesis (Goldman et al., 1999; Kagan et al., 1999). In effect, the reactivity of phenoxyl radicals determines, to a large extent, their overall cytotoxicity and genotoxicity in MPO-expressing CD34+ cells, the likely precursors from which t-AML arises. Hence, characterizing the interactions of etoposide phenoxyl radicals with major cellular components is essential for a better understanding of this drug's effects on cells (Kagan et al., 1999, 2001).
The most direct way to detect and monitor the free radical MPO-initiated reaction is via EPR spectroscopy. We reported previously that EPR detection of a phenoxyl radical of etoposide is feasible in MPO-rich human myeloid leukemia HL60 cells (Kagan et al., 2001). EPR detection of the radicals became possible after depletion of GSH and other thiols, suggesting that etoposide radicals (etoposide-O•) displayed reactivity toward these abundant intracellular reductants (Kagan et al., 1999). Furthermore, possible involvement of secondary reactions of thiol radicals leading to the production of superoxide radicals and other reactive oxygen species were considered as important cytotoxic and genotoxic events (Kagan et al., 1999, 2001).
To further evaluate whether MPO is a cellular determinant of etoposide oxidation, genotoxicity, and leukemogenesis, we evaluated MPO-catalyzed production of etoposide phenoxyl radicals in growth factor-mobilized human CD34+ cells, a proximal progenitor model for t-AML. We report for the first time the detection of the EPR signal of etoposide phenoxyl radicals in intact CD34+ cells and demonstrate that this process is MPO-dependent and leads to the depletion of intracellular thiols. In addition, our results demonstrate an MPO-dependent component of etoposide-induced DNA damage and MLL gene rearrangements, providing “proof-of-principle” evidence for MPO as a determinant of etoposide leukemogenesis.
Materials and Methods
Materials.
Etoposide (VP-16), phenol, hydrogen peroxide, succinylacetone (SA), guaiacol, 3-amino-1,2,4-triazole (3-AT), phenylmethylsulfonyl fluoride, glucose, cetylmethylammonium bromide, glucose, HEPES, dimethyl sulfoxide (DMSO), sodium chloride, sodium phosphate, diethylenetriaminepentaacetic acid (DTPA), and myeloperoxidase (from human leukocytes, EC 1.11.1.7) were purchased from Sigma-Aldrich (St. Louis, MO). Triton X-100 (t-octylphenoxy polyethoxyethanol) was from Bio-Rad Laboratories (Hercules, CA). ThioGlo-1 was from Covalent Associates, Inc. (Woburn, MA).
Cell Culture and CD34+ Cell Isolation.
Human umbilical cord blood (CB) samples were obtained immediately after delivery in accordance with institutional guidelines and placed in 50-ml tubes containing anticoagulant citrate dextrose solution (ACD-A; Cytosol Labs, Braintree, MA). The CB was diluted with calcium- and magnesium-free phosphate-buffered saline (PBS) (+) 0.6% ACD-A, and low-density mononuclear cells were isolated by Ficoll-Paque density gradient centrifugation for 30 min at 400g (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). CB mononuclear cells were washed twice in PBS and resuspended in PBS plus 0.6% ACD-A for magnetic labeling and separation. CD34+ progenitor cells were isolated using immunomagnetic selection techniques. In brief, cells were incubated with blocking reagent (human IgG) and QBEND/10 CD34 antibody for 15 min at 4°C and washed in PBS/ACD-A followed by incubation with a secondary antibody-magnetic microbead conjugate for an additional 15 min at 4°C. The unlabeled fraction of CD34(−) cells were separated from the labeled CD34+ fraction on a high-gradient magnetic separation column (Miltenyi Biotec, Sunnyvale, CA). Isolated CD34+ cells were grown in 95% humidity under 5% CO2 in air at 37°C for up to 2 weeks in Iscove's modified Dulbecco's minimal essential medium, supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 ng/ml each of interleukin-3, stem cell factor, and granulocyte colony-stimulating factor. Human myeloid HL-60 cells (from the American Type Culture Collection, Manassas, VA) were grown continuously in RPMI 1640 medium supplemented with 15% FBS and 2 mM l-glutamine. CD34+ and HL-60 cells were collected for experiments in mid-log phase growth (5–8 × 105 cell/ml).
MPO Peroxidase Activity in Cell Homogenates.
CD34+ cells were harvested by centrifugation at 500g for 5 min. Pellets were washed twice with buffer A containing 25 mM HEPES and 5 mM NaH2PO4, pH 7.4, 10 mM glucose, 115 mM NaCl, 5 mM KCl, and 1 mM MgCl2. Cell homogenates were prepared by freezing at −80°C and then thawing cells. Measurements of MPO activity were made at room temperature using guaiacol as substrate. Guaiacol oxidation was monitored by changes of absorbance at 470 nm (ε = 26.6 mM−1 · cm−1) using a spectrophotometer (Shimadzu, Kyoto, Japan). Cell homogenates (0.1–0.4 × 106 cells) were added to 50 mM disodium phosphate buffer containing 0.1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, 10 mM guaiacol, 0.02% cetylmethylammonium bromide, and 3.75 mM 3-AT, pH 7.0. The reaction was started by the addition of 500 μM H2O2. Activity of MPO was calculated as nanomoles of tetraguaiacol formed per minute per 106 cells.
In Vitro MPO Peroxidase Activity Using Guaiacol and Etoposide as Substrates.
Peroxidase activity of purified myeloperoxidase was assayed both with guaiacol and etoposide in 50 mM sodium phosphate buffer, pH 7.4, and 100 μM DTPA at 25°C. Kinetics of guaiacol oxidation was monitored at 470 nM (ε = 26.6 mM−1 · cm−1) after the addition of 100 μM H2O2 to the solution of 10 nM MPO and 500 μM guaiacol. Oxidation of etoposide (200 μM) by MPO (100 nM) was detected by measurement of the EPR spectra of etoposide phenoxyl radicals 1 min after the addition of 100 μM H2O2 to the solution. The concentration of etoposide radicals produced during the course of the MPO reaction was estimated by the use of stable nitroxide 4-amino-2,2,6,6-tetramethylpiperidine-1-oxyl as a standard. Double integration of the spectra was performed using the WinSim Inc. Process simulation program (Laboratory of Molecular Biophysics, National Institute of Environmental Health Sciences, Bethesda, MD). The relative activity of guaiacol and etoposide as MPO substrates was characterized by calculating the number of substrate molecules converted to product per unit time [kcat (micromoles of substrate per minute per micromole of enzyme].
Immunoblotting for MPO.
Whole-cell lysates were prepared from 1 million pelleted CD34+ cells by the addition of SDS-polyacrylamide gel electrophoresis sample buffer [50 mM Tris-HCl, pH 6.8, 1% (w/v) SDS, 10% (v/v) glycerol, and 0.5% (v/v) 2-mercaptoethanol], followed by boiling for 5 min and brief sonication. Protein samples of CD34+ cell lysate (15 μg) were resolved using 10% (w/v) SDS-polyacrylamide gel electrophoresis and then transferred to nitrocellulose. Visual inspection of Ponceau S-stained nitrocellulose membranes was used to ensure equivalent loading/transfer of the lysates. Membranes were blocked with nonfat dry milk [3% (w/v)] in PBS containing 0.05% (w/v) Tween 20 and then incubated with 1:40,000 dilutions of primary rabbit anti-MPO antibodies kindly provided by Dr. William Nauseef (University of Iowa School of Medicine, Iowa City, IA). The secondary donkey anti-rabbit antibody used at 1:20,000 dilution was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Bound antibody was detected using enhanced chemiluminescence (PerkinElmer Life and Analytical Sciences, Waltham, MA).
Samples for EPR Measurements.
In experiments with purified MPO, etoposide (200 μM) was added to the solution of MPO (25 nM) in phosphate buffer (50 mM and 100 μM DTPA, pH 7.4 at 25°C). Etoposide radical formation was monitored at room temperature 1 min after the addition of 100 μM H2O2. Generation of these radicals in suspensions of HL-60 cells (3 × 106 cell/ml) or CD34+ cells (8–10 × 106 cell/ml) in buffer A were recorded by EPR spectroscopy at room temperature. 3-AT (6 mM) was added to the cell suspension and incubated for 6 min, after which etoposide (200 μM) was added. Incubation for 2 min with etoposide was followed by the addition of H2O2 (100 μM), and EPR spectra were recorded beginning 1 min thereafter. For thiol depletion, the cells were preincubated in buffer A for 10 min with ThioGlo-1 (30 μM) followed by washout of this maleimide reagent and additions sequentially of 3-AT, etoposide, and H2O2 as indicated above. In experiments with phenol, the experimental procedure was identical except that phenol (at various concentrations) was added 2 min after 3-AT.
EPR Measurements.
EPR spectra were recorded on a JEOL-REIX spectrometer with 100-kHz modulation (JEOL, Kyoto, Japan) in gas-permeable Teflon tubing (0.8 mm internal diameter, 0.013 mm thickness) obtained from Alpha Wire Corporation (Elizabeth, NJ). The tubing was filled with 60 μl of sample, folded doubly, and placed in an open 3.0 mm internal diameter EPR quartz tube. Etoposide phenoxyl radical spectra were recorded at 3350 G, center field; 50 G, sweep width; 10 mW, microwave power; 0.5 G, field modulation; 103, receiver gain; 0.1 s, time constant; and 2 min, scan time. The time course of etoposide radical EPR signals was obtained by repeated scanning (25 s) of part of the spectrum (3350 G, centered field; 5 G, sweep width; and other instrumental conditions were the same). A computer simulation of the experimental spectrum was made by the use of WinSim software package, and the numbers of hyperfine couplings for etoposide phenoxyl radicals were published previously (Kalyanaraman et al., 1989).
Flow Cytometry Assay for Intracellular Thiols in Native Cells.
After treatment, the cells were collected and resuspended in PBS at a density of 0.1 × 106 cells/ml followed by incubation with ThioGlo-1 (10 μM) at room temperature for 10 min after washing once with PBS. The fluorescence of ThioGlo-1 inside cells was measured using a FACScan (BD Biosciences, San Jose, CA) flow cytometer, equipped with a 488-nm argon ion laser and supplied with the Cell Quest software. Mean fluorescence intensity from 104 cells was acquired using a 530-nm filter (FL-1 channel). Although excitation of ThioGlo-1 at 488-nm is at the tailing end of the absorption spectrum, absorbance was sufficient for the recording of emission spectra.
Fluorescence Assay for Low Molecular Weight Thiols (GSH).
Low molecular weight thiols (predominantly GSH) in cells were determined using ThioGlo-1, which produces a highly fluorescent adduct upon its reaction with SH-groups. Cells (0.2 × 105) were suspended in PBS and lysed by repeated freeze-thaw. GSH content was estimated by an immediate fluorescence response registered upon the addition of ThioGlo-1 to the cell homogenate using excitation at 388 nm and emission at 500 nm. The total amount of protein was determined using the Bradford assay.
Cytogenetic Analysis for MLL Gene Rearrangements.
Growth factor-mobilized CD34+ cells were treated for 60 min with etoposide (50 μM) or vehicle (DMSO). Cells were then washed free of drug and allowed to grow for an additional 7 days, after which they were treated with demecolcine (Colcemid, 0.1 mg/ml; Invitrogen, Carlsbad, CA) for 1 h before standard cytogenetic harvesting. After mitotic arrest, the cells were incubated in 0.075 M KCl, fixed with Carnoy's fixative (3:1 methanol/glacial acetic acid), and slides were prepared for cytogenetic analysis. A dual-color break-apart DNA probe for the detection of human MLL gene rearrangements was obtained from Abbott Molecular Inc. (Des Plaines, IL). This probe consists of a 350-kilobase segment centromeric to the MLL breakpoint cluster region (bcr) labeled in SpectrumGreen and a 190-kilobase segment, mostly telomeric to the MLL bcr, labeled in SpectrumOrange. The probe was diluted 1:5 in tdenhyb (Insitus Biotechnologies, Albuquerque, NM). Fluorescence in situ hybridization (FISH) assays were carried out to detect and quantify MLL gene rearrangements. Slides were pretreated with RNase, dehydrated in an ethanol series, denatured in 70% formamide, and hybridized with probe overnight at 37°C in a humidified chamber. Posthybridization washes were carried out according to the Abbott Molecular protocol. The slides were stained with 4′,6-diamidino-2-phenylindole and mounted with antifade composed of 1 mg/ml 1,4-phenyene-diamine (Sigma-Aldrich) in 86% glycerol/PBS at pH 8.0. FISH signals on metaphase spreads and in interphase nuclei were analyzed. Hybridizations enabled analysis of between 124 and 274 cells for DMSO and between 213 and 220 cells for the etoposide-treated samples. A yellow (orange + green) fluorochrome fusion signal suggested the presence of an intact MLL gene, whereas separation of the two signals indicated the presence of an MLL gene rearrangement. All FISH analyses were carried out using an Olympus BX61 epifluorescence microscope (Olympus Microscopes, Melville, KY). The Genus software platform on the Cytovision System was used for image capture and analysis (Applied Imaging, San Jose, CA).
Comet Assays.
The alkaline single-cell gel electrophoresis assay for DNA damage (Comet assay) was performed essentially according to instructions provided in the Trevigen Comet Assay kit (Trevigen, Gaithersburg, MD). CD34+ cells growing in complete Iscove's DMEM supplemented with FBS, and growth factors were incubated in the absence or presence of succinylacetone for 63 h, after which cells were washed and resuspended in buffer A at 37°C to a density of approximately 500,000 cells/ml. Cells were incubated for 30 min with etoposide (5 μM) dissolved in DMSO or with DMSO vehicle alone [0.2% (v/v)] followed by centrifugal collection of cell pellets. Gentle resuspension in 1.5 ml of ice-cold 1× PBS (calcium- and magnesium-free) was followed by a second centrifugation and resuspension of pellets in 0.5 ml of the same ice-cold PBS. Thirty microliters of cell suspension was carefully mixed with 300 μl of low melting temperature agarose (Trevigen LMAgarose) at 39°C. A cell/agarose mixture (75 μl) was then transferred evenly to Trevigen COMET glass slides, which were immediately placed in a desiccator jar at 4°C in the dark for 30 min to allow the agarose to set. Slides were then transferred into prechilled Trevigen lysis solution for 2 h in the dark. Next, the slides were immersed in an alkaline solution (pH > 13, 300 mM NaOH, and 1 mM EDTA) for 1 h in the dark at room temperature. Slides were transferred to the center of a horizontal gel electrophoresis apparatus (with electrodes 34 cm apart) containing just enough ice-cold alkaline solution (pH >13, 300 mM NaOH) to cover the slides. Electrophoresis proceeded in a cold room at 1 V/cm for 45 min. After electrophoresis, the slides were drained of excess alkaline solution, immersed in 70% ethanol for 5 min, and air-dried overnight. Slides were stained with SYBR green, and images were visualized under a fluorescence microscope and captured with a charge-coupled device camera. Images were imported and analyzed using a version of the Comet Assay Software Project (CASP) the public domain program specifically designed for the Comet assay (Konca et al., 2003). From each slide, at least 150 cells were analyzed. DNA damage is presented as the Olive tail moment in etoposide-treated minus DMSO-treated cells. The Olive tail moment is defined as the product of the comet tail length and the fraction of total DNA in the comet tail (Olive, 2002).
Statistical Analysis.
The results are presented as the mean values ± S.D. values for n ≥ 3, and statistical analysis was performed by one-way analysis of variance. The statistical significance of differences was set at P < 0.05.
Results
Detection of the Etoposide-O• in Intact CD34+ Cells.
Our previous work demonstrated MPO-dependent one-electron oxidation of etoposide to its phenoxyl radical, etoposide-O•, in intact HL-60 leukemia cells (Kagan et al., 2001). In those studies etoposide-O• was directly detectable by EPR spectroscopy after depletion of nonprotein thiols with the maleimide reagent ThioGlo-1. Using similar experimental conditions, we now detect the characteristic EPR signal for etoposide-O• with g = 2.004 in growth factor-mobilized human CD34+ myeloid progenitor cells isolated from umbilical cord blood (Fig. 1A, b). Computer simulation of the experimental spectrum provided additional proof that the radical was etoposide-O• (Fig. 1A, c). For our experimental conditions the best simulation of the spectrum was achieved with hyperfine couplings aOCH3H = 1.4 G (6), aringH = 1.4 G (2), aβH = 4.3 G (1), and aγH = 0.64 G (1). The numbers in parentheses denote the number of equivalent protons. These are in good agreement with hyperfine couplings published earlier for the etoposide phenoxyl radical (Kalyanaraman et al., 1989). Growth factor mobilization of the CD34+ progenitors leads to a progressive expression of MPO (Morabito et al., 2005). MPO activity in these CD34+ cells cultured for 1 week in the presence of 100 μg/ml stem cell factor, interleukin-3, and granulocyte colony-stimulating factor was found to be 9.0 ± 2.5 nmol tetraguaiacol formed/min/106 cells. In contrast, freshly isolated CD34+ cells contained no detectable MPO activity and did not convert etoposide to its phenoxyl radical (Fig. 1A, a). Together our results indicate that etoposide-O• is formed in myeloid CD34+ cells and that this oxidation of etoposide is dependent on the level of MPO.
EPR signal of etoposide phenoxyl radicals (A) and MPO levels (B) in CD34+ cells. A, CD34+ cells (8–10 × 106 cells/ml) pretreated with ThioGlo-1 were incubated with 6 mM 3-AT for 6 min. Etoposide (200 μM) was then added, and cells were incubated 2 min longer. EPR signal of etoposide-O• radical was recorded beginning 2 min after the addition of 100 μM H2O2. a, freshly obtained CD34+ cells; b, growth factor-mobilized CD34+ cells; c, computer simulation of experimental EPR spectrum of etoposide phenoxyl radical. The computer simulation was obtained by using a line width of 0.2 G and 50% Lorentzian/50% Gaussian line shapes. d, mobilized CD34+ cells without addition of H2O2; e, mobilized CD34+ cells incubated for 63 h in the presence of succinylacetone (200 μM), an inhibitor of heme synthesis; f, mobilized CD34+ cells in the presence of cyanide (0.5 mM). B, Western blot analysis of MPO levels in CD34+ cells without (−SA) or with (+SA) a 63-h exposure to 200 μM succinylacetone, a heme synthesis inhibitor. The level of mature, enzymatically active MPO (59 kDa) was decreased in the SA-treated compared with untreated CD34+ cells.
Because both etoposide and the added MPO cosubstrate H2O2 are cytotoxic, thereby releasing cellular contents (including MPO), we next established that etoposide-O• radicals were generated exclusively inside cells. CD34+ cells pretreated with ThioGlo-1 were incubated with 200 μM etoposide and 100 μM H2O2 for 45 min (H2O2 was added at time 0 and every 15 min thereafter). Viability of cells treated with etoposide and H2O2 was 91 ± 5% (by trypan blue dye exclusion assay) after 45-min incubation. Peroxidase activity in collected supernatants during this incubation period was virtually undetectable. In addition, the magnitude of the EPR signal for etoposide-O• in supernatants collected at 15 and 45 min (after fresh addition of etoposide and H2O2)was 5 to 10% of that observed in intact cells (results not shown). Together, these results indicate that etoposide-O• formation is an intracellular event and that CD34+ cells remained intact during the incubation period with H2O2 and etoposide.
Formation of Etoposide-O• in CD34+ Cells Is MPO-Dependent.
To confirm the involvement of endogenous MPO in one-electron oxidation of etoposide in CD34+ cells, we compared the main characteristics of the peroxidase reaction and etoposide-O• radical generation. First, as indicated above, the magnitude of the EPR signal of etoposide radicals correlated with the MPO activity in freshly obtained CD34+ cells compared with cells grown for 1 week in growth factor containing media (Fig. 1A, a compared with b). In addition, the EPR signal of etoposide-O• in CD34+ cells was not detected in the absence of the MPO cosubstrate, H2O2 (Fig. 1A, d).
We next demonstrated that the formation of etoposide-O• radicals in CD34+ cells is a heme-mediated process. To this end, we used SA, an inhibitor of heme synthesis (Pinnix et al., 1994) and studied its effect on the EPR signal of etoposide-O• radicals in mobilized CD34+ cells. Immunoblot analysis (Fig. 1B) indicated that mobilized CD34+ cells contain mature (heavy subunit) MPO. A 48-h incubation with 200 μM SA dramatically decreased levels of MPO in CD34+ cells (Fig. 1B). Quantitation of replicate immunoblots indicated a reduction of MPO levels to 33.0 ± 5.5% of control cells. Under these conditions (+SA), MPO activity was diminished to 30.5 ± 7.1% of that seen in control cells (data not shown). Correlating with a decrease in the level and activity of MPO, formation of etoposide-O• was dramatically decreased in these SA-treated heme-depleted cells. The magnitudes of the EPR signal for etoposide-O• were 45 ± 8 AU and 13 ± 4 AU for control CD34+ cells and for SA-treated CD34+ cells, respectively (n = 3). The representative spectra are shown in Fig. 1A, b and e. Cyanide (0.5 mM), an inhibitor of heme-containing enzymes, inhibited etoposide-O• formation by approximately 90% (Fig. 1A, f). Although cyanide and SA are not highly specific inhibitors of MPO but rather block heme-containing enzymes and total heme synthesis, respectively, together, these results strongly suggest that H2O2-dependent MPO catalysis is involved in the generation of etoposide-O• radicals, especially because MPO is a major heme-containing protein in growth factor-mobilized CD34+ cells and there is little expression of cytochromes P450 in CD34+ cells capable of etoposide oxidation (Soucek et al., 2005).
Effect of Phenol on MPO-Dependent Etoposide-O• Formation.
The high-oxidizing potential of the reactive intermediate MPO compound I (1.35 V) allows for one-electron oxidation of reducing substrates such as phenolic compounds to form phenoxyl radicals (Jantschko et al., 2005; Davies et al., 2008). Despite its low redox potential (E0 = 0.56 V), etoposide is expected to be a very poor substrate for MPO because access of this large phenolic compound (molecular weight = 589) to the MPO active site is highly constrained. The MPO active site is located at the base of a narrow and deep heme pocket (Day et al., 1999; Zhang et al., 2002). We measured MPO activity using both etoposide and guaiacol as substrates. MPO activity toward etoposide was 2 orders of magnitude less than that toward guaiacol (kcat = 8.8 ± 2.4 versus 1050 ± 150/min, respectively). This result is consistent with our observation that high concentrations of etoposide are required to observe MPO-catalyzed production of etoposide-O•.
Compared with etoposide, small phenolic molecules (phenol, tyrosine) are good substrates of MPO (Goldman et al., 1999). These small molecules may act as cosubstrates for oxidation of large, bulky compounds such as etoposide because they have free access to the MPO active site and because of the relatively high oxidizing potential (approximately 0.7–0.9 V) of their phenoxyl radicals compared with etoposide. Hence, to further indicate that MPO is responsible for H2O2-dependent etoposide oxidation in CD34+ cells, we examined a specific feature of MPO catalysis, namely the amplification of MPO-induced oxidation of the bulky etoposide molecule in the presence of the smaller phenol molecule. Phenol-derived phenoxyl radicals were not detected under our experimental conditions. These phenoxyl radicals are short-lived because of their high oxidizing potential and chemical reactivity. Phenol-derived phenoxyl radicals recombine with a second order rate constant of approximately 108 M−1 · s−1 (Goldman et al., 1997) compared with the recombination rate constant for the relatively long-lived etoposide phenoxyl radicals; 3 × 103 M−1 · s−1 (Tyurina et al., 2006).
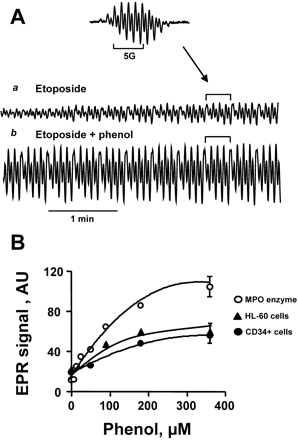
Figure 2A shows the effect of phenol on the H2O2-dependent formation of etoposide-O• by purified MPO. Given the second order recombination rate constant for etoposide radicals of 3 × 103 M−1 · s−1 (Tyurina et al., 2006), their lifetime is expected to exceed the time of measurements (approximately 250 s) under our experimental conditions. Hence, progressive accumulation of etoposide radicals can be detected. We easily measured the time-dependent increase of EPR signals of etoposide-O• when etoposide and H2O2 were added to purified MPO (Fig. 2Aa). Under the same experimental conditions but in the presence of 100 μM phenol, the production of etoposide-O• after 1 min increased dramatically (Fig. 2Ab).
Phenol-induced amplification of etoposide radical formation by MPO. A, EPR spectrum (inset) and kinetics of etoposide-O• generated by the addition of purified MPO. MPO (25 nM) was added together with etoposide (200 μM) (a) or with etoposide and phenol (100 μM) (b). The time course of the EPR signal was monitored beginning 1 min after the addition of H2O2 (100 μM). B, magnitude of EPR signal of etoposide-O• 1 min after the addition of H2O2 to a solution containing purified MPO (○, top curve) or suspensions of cells in buffer A [▴, HL60 cells; ●, growth factor-mobilized CD34+ cells] in the presence of various concentrations of phenol. HL60 cells treated with ThioGlo-1 were at a concentration of 3 ± 0.5 × 106 cells/ml. All other experimental conditions were the same as detailed in the legend to Fig. 1.
The concentration-dependent effects of phenol on purified MPO enzyme oxidation of etoposide are demonstrated in Fig. 2B, top. If cellular etoposide oxidation is similarly catalyzed by MPO, then phenol-induced enhancement of etoposide oxidation should be observed in intact CD34+ cells. Indeed, we next demonstrate the phenol-dependent amplification of the EPR signal of etoposide phenoxyl radicals in MPO-rich human leukemia HL-60 cells and in growth factor-mobilized CD34+ cells (Fig. 2B). In cells, there are many potential intracellular targets beside etoposide and thiols for the highly reactive phenol radicals. For example, in contrast to etoposide phenoxyl radicals, phenol-derived phenoxyl radicals react with lipids and proteins and oxidize them very well (Goldman et al., 1999; Tyurina et al., 2006). This may account for the less pronounced phenol potentiation of etoposide-O• production in cells compared with an in vitro isolated enzyme system (Fig. 2B). Using this in vitro model system, phenol at 10 μM was sufficient to increase the etoposide-O• signal more than 2-fold. In cells, by comparison, reliable increases of the etoposide radical signal (30%) were observed at much higher added phenol concentrations (50 μM) and only after depletion of intracellular thiols (using ThioGlo-1), one of the main targets of phenoxyl radicals (Fig. 2B). On the other hand, it is also possible that the increase of etoposide radical signals under these conditions may be mediated via secondary phenoxyl radical reactions such as oxidation of tyrosines, tryptophans, and lipids, resulting in intermediates with high oxidizing redox potentials. Overall, in cells, the phenol-dependent enhancement of etoposide-O• production indicates that MPO is involved in etoposide oxidation.
Effects of Etoposide on H2O2-Induced Oxidation of Endogenous Thiols in Human CD34+ Cells.
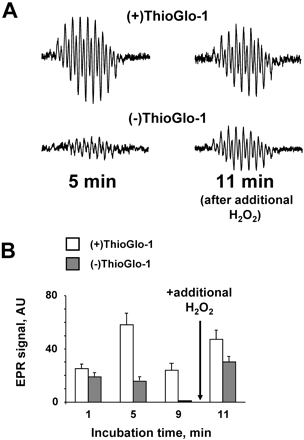
Our previous studies demonstrated that etoposide-O• radicals are reactive toward intracellular reductants such as ascorbate (Kagan et al., 1994, 1999) (suggesting a future chemoprevention strategy to diminish the leukemogenic effects of etoposide) and endogenous thiols such as GSH (Tyurina et al., 1995; Kagan et al., 1999, 2001). The addition of ThioGlo-1, a maleimide reagent capable of titrating out thiols, was essential for observation of intracellular etoposide-O• in human myeloid leukemia HL-60 cells (Kagan et al., 2001). To determine whether titrating out thiols is similarly required to observe etoposide-O• in CD34+ cells, we compared the time course of etoposide-O• formation in this cell population before and after treatment with ThioGlo-1. We found that the maximal EPR signal for etoposide-O• was several times greater after ThioGlo-1 treatment (Fig. 3A). When cells were pretreated with Thioglo-1, the magnitude of the signal increased over time and then decreased (Fig. 3B), probably because of the depletion of H2O2. In the absence of ThioGlo-1, there was a less pronounced increase in the EPR signal and a more dramatic decrease of the signal within 10 min of the addition of H2O2 (Fig. 3B). Another addition of H2O2 (100 μM) after 10 min reconstituted the EPR signal for etoposide-O• observed 1 min later with a greater signal recorded in cells pretreated with ThioGlo-1 (Fig. 3B).
Time course of etoposide-O•formation in CD34+ cells. A, EPR spectra of etoposide radical formation in growth factor mobilized CD34+ cells treated with (top spectra) or without (bottom spectra) ThioGlo-1. CD34+ cells (9–11 × 106 cells/ml) in buffer A were incubated with 6 mM 3-AT, etoposide (200 μM), and H2O2 (100 μM) was added sequentially as in Fig. 1. Complete spectra were subsequently recorded 5 min after the addition of 100 μM H2O2. After 10 min, an additional aliquot of H2O2 was added, and spectra were recorded 1 min later. B, magnitude of EPR signal of etoposide-O• in suspensions of CD34+ cells treated with (□) or without (■) ThioGlo-1 at different incubation times with 200 μM etoposide and 100 μM H2O2.
Repeated additions of H2O2 and etoposide/H2O2 in cell suspensions not pretreated with Thioglo-1 resulted in a progressive increase in the peak magnitude of etoposide-O• radical formation followed by a decrease in EPR signal (Fig. 4A). These results suggest that etoposide-O• reactivity toward endogenous thiols caused oxidation and depletion of the thiol pool, allowing for an initial increase of the signal upon further addition of H2O2 and etoposide/H2O2 (Fig. 4A). Hence, reactivity of etoposide-O• toward intracellular thiols should be observable directly through oxidation and depletion of SH-groups. Therefore, we measured low molecular weight thiols and protein thiols in growth factor-mobilized CD34+ cells after incubation with etoposide and H2O2 (Fig. 4B). At various time points, aliquots of CD34+ cell suspension were taken for measurements of low molecular weight thiol content by flow cytometry after addition of ThioGlo-1. In addition, for estimation of low molecular weight thiol content in CD34+ cells, the immediate fluorescence response to the addition of ThioGlo-1 in cell homogenates was measured in a spectrofluorometer. Protein SH-groups were determined by the additional increase in fluorescence response after the addition of SDS (4 mM) to the same cell homogenates. The validity of this technique for assessment of low molecular weight thiols and protein-SH groups has been demonstrated previously (Kagan et al., 2001).
Oxidation of SH-groups and radical formation in the presence of etoposide in CD34+ cells. A, time course of etoposide radical formation in the suspension of growth factor mobilized CD34+ cells (without ThioGlo-1 treatment) with repeated addition of H2O2. Etoposide (100 μM) and H2O2 (100 μM) were added to cell suspensions at time 0. As indicated, more H2O2 (100 μM) and etoposide (100 μM) plus H2O2 (100 μM)) were added to cell suspensions 10 and 30 min later, respectively. Samples for the measurements of thiols were taken before any additions (control) and then 30 and 45 min after first addition of H2O2. B, free thiol group content in CD34+ cells was measured by addition of ThioGlo-1 after cell incubation with etoposide and H2O2 either to cell suspensions (a) or to cell lysates (b and c). a, the fluorescence of ThioGlo-1 within intact CD34+ cells assessed by flow cytometry. (Inset indicates the decrease in total free thiols groups in cells 45 min after several additions of etoposide and H2O2 as indicated in A); b, time-dependent change in the level of low molecular weight thiols (predominantly GSH) in cell lysates; c, time-dependent change in the level of free protein thiols in cell lysates. *, P < 0.05 compared with 0 time control.
We observed a time-dependent decrease in low molecular weight thiol content both in intact cells by flow cytometry (Fig. 4Ba) and in cell homogenates by spectrofluorometry (Fig. 4Bb). In contrast, there was no statistically significant decrease in free total protein SH content after 45-min incubation with etoposide and H2O2 (Fig. 4Bc).
These results are consistent with the idea that the abundant low molecular weight thiols such as GSH are the immediate reductants of the etoposide-O•, whereas protein SH-groups are oxidized only after depletion of endogenous GSH or when GSH levels are relatively low. In support of this concept, we have demonstrated previously in MPO-rich HL-60 cells that pretreatment with ThioGlo-1 to deplete cells of GSH resulted in enhanced oxidation of protein thiols (Kagan et al., 2001). When GSH levels are low, etoposide-mediated protein SH oxidation in CD34+ cells may be significant at the level of DNA topoisomerase II cysteines as a determinant of cytotoxicity, genotoxicity, and the known leukemogenicity of this agent.
Effects of Etoposide on DNA Damage and MLL Gene Rearrangement in CD34+ Cells: Role of Myeloperoxidase.
When CD34+ cells were pretreated with 200 μM SA for 63 h to decrease MPO levels, etoposide (5 μM)-induced DNA damage assessed by Comet assay (after 30 min) was significantly reduced compared with CD34+ cells with replete MPO (Fig. 5). DNA damage was quantified in etoposide-treated compared with vehicle alone (DMSO) controls in SA pretreated or untreated cells. SA did not perturb vehicle alone control levels of DNA damage (data not shown). These results indicate an MPO-dependent component of etoposide-induced DNA damage in CD34+ cells consistent with the idea that MPO catalyzed oxidation of etoposide can result in increased genotoxicity and potentially carcinogenicity. To further establish the role of MPO in etoposide-mediated carcinogenicity, we examined the effects of SA pretreatment on etoposide-induced MLL gene rearrangements in CD34+ cells. Growth factor-mobilized human CD34+ cells were pretreated for 48 h with or without SA (200 μM) to deplete cells of mature MPO. Cells were then incubated for 60 min with 50 μM etoposide (VP-16). Cells were washed free of drug and allowed to grow for an additional 7 days, after which metaphase chromosomes were analyzed for MLL gene rearrangements (MLLR) by FISH using a dual-color break-apart probe for MLL. For each etoposide treatment, more than 200 cells were analyzed. No MLLR were observed in controls. For etoposide-treated CD34+ cells 3.8% of cells exhibited MLLR, and this was reduced to 0.5% when cells were depleted of MPO. Hence, the depletion of MPO resulted not only in decreased etoposide-O• formation (Fig. 1A, e) but also in a reduction in both etoposide-induced DNA damage (Fig. 5) and MLLR.
MPO-dependent etoposide-induced DNA damage in CD34+ cells. Growth factor-mobilized CD34+ cells were incubated for 63 h in the absence or presence of SA (200 μM) followed by a 30-min incubation with etoposide (5 μM) or vehicle control [DMSO 0.2% (v/v)]. Cells were then processed and evaluated for DNA damage by Comet assay as described under Materials and Methods. Results shown are the mean ± S.E.M. for triplicate measurements during each of three experiments run on separate days. *, P < 0.05 comparing etoposide effects in SA-treated and untreated cells.
Discussion
This report presents, for the first time, direct detection and monitoring of etoposide radicals and their enhancement by small phenolic molecules in CD34+ myeloid progenitor cells. Detection of free radicals in cells is not trivial. Few reports have been published demonstrating endogenous production of free radicals in cells or in animals. An ESR spin-trapping system was successfully implemented to detect free radicals produced in rodents after treatment with tert-butyl hydroperoxide (Hix et al., 2000). A spin-trapping technique was used as well for the detection of radicals inside macrophages (Lopes de Menezes and Augusto, 2001). We presented evidence previously for the formation of etoposide-O• in HL-60 human myeloid leukemia cells (Kagan et al., 2001) using clinically relevant concentrations of etoposide. To date, no reports have been published demonstrating the direct detection of MPO-derived radicals in normal cells.
Detection of etoposide phenoxyl radicals in bone marrow CD34+ progenitor cells, the likely precursors from which myeloid tumors arise, is critical to understand the genotoxic effects of etoposide. These myeloid progenitor cells contain MPO even in the early stages of maturation (Strobl et al., 1993). In our experiments, after the addition of etoposide to human CD34+ cells, we were able to detect and monitor the EPR signal of etoposide phenoxyl radicals. Detection of etoposide phenoxyl radicals was dependent on the depletion of nonprotein thiols such as GSH by preincubation of cells with the maleimide reagent ThioGlo-1. At the same time, we were able to obtain EPR spectra with distinguishable EPR signals even without ThioGlo-1. The radical signal can be measured reliably after repeated additions of H2O2 and (etoposide + H2O2), indicating that intracellular reactions of etoposide radicals lead to the depletion of endogenous GSH (Figs. 3 and 4A). Because the sensitivity of EPR spectroscopy is relatively low, we used higher concentrations of etoposide. Nevertheless these concentrations were clinically relevant because etoposide plasma levels of 100 μg/ml (180 μM) can be achieved in patients receiving high doses of this drug (Stremetzne et al., 1997). Under our experimental conditions, the concentration of etoposide phenoxyl radicals accumulated inside CD34+ cells was as high as 0.2 ± 0.05 μM etoposide-O•/106 cells.
We provide several lines of evidence associating the observed etoposide-O• radical signals in CD34+ cells with the activity of the heme protein, MPO. First, peroxidase activity correlated with the detection of the EPR signal of etoposide-O•. Second, H2O2, an MPO cosubstrate, dramatically stimulated formation of the etoposide phenoxyl radical. Third, generation of etoposide phenoxyl radicals was demonstrated to be a heme-mediated process because phenoxyl radical formation was inhibited by the addition of cyanide or by preincubation of cells with the heme-synthesis inhibitor, succinylacetone. Finally, the established MPO substrate, phenol, amplified production of etoposide phenoxyl radicals within cells, strongly suggesting that MPO-catalyzed one-electron oxidation is responsible for etoposide “activation.”
As a bulky molecule, etoposide is a relatively poor substrate for MPO compared with the small molecule phenol, which has greater access to the relatively restricted active site of the enzyme (Day et al., 1999; Zhang et al., 2002). MPO-dependent production of nonspecific phenol radicals results in subsequent production of the etoposide-O• based on its specific property as a phenoxyl radical with low redox potential compared with phenol-derived phenoxyl radicals (Goldman et al., 1997; Goldman et al., 1999). Hence, incubation of MPO-rich cells with both phenol and etoposide may be responsible for converting nonspecific phenol radicals, which are broadly reactive toward different biomolecules, to specific interactions of etoposide-O• radicals with GSH and protein-SH groups. This specificity may be responsible for modified and/or inhibited protein activities.
Because the etoposide phenoxyl radical is relatively long-lived compared with the phenol radical (Goldman et al., 1997; Tyurina et al., 2006), the oxidative potential of MPO can be more readily transferred to the nucleus when cells are relatively depleted of intracellular reductants. In addition, it has been demonstrated that MPO can be found in the nucleus of normal and leukemic human myeloid cells (Murao et al., 1988). Hence, one suggestion from our results is that exposure of MPO-containing myeloid progenitors to etoposide or concurrently to etoposide and phenols increases the risk of etoposide-induced secondary acute myeloid leukemias based on increased oxidative stress and potential oxidative DNA damage, resulting in abasic DNA sites. These abasic sites can act to enhance DNA topoisomerase II poisoning (Kingma et al., 1997) and may thereby increase the recombinogenic activity of etoposide.
In support of the idea that hematopoietic cell cytotoxicity (and presumably genotoxicity) can be enhanced by MPO-catalyzed oxidation of phenol, it was reported that phenol stimulated hydroquinone oxidation by peroxidases (Smith et al., 1989). In addition, repeated coadministration of phenol and hydroquinone to B6C3F1 mice resulted in a dramatic decrease in bone marrow cellularity (Eastmond et al., 1987). Together with the results presented here, these reports raise a cautionary note concerning environmental phenol exposure in patients receiving etoposide therapy. In contrast, dietary flavonoids, relatively weak oxidants, can be viewed as potentially important competitive inhibitors of MPO-catalyzed etoposide oxidation useful for the prevention of etoposide genotoxicity.
Peroxidase-dependent formation of phenoxyl radicals in the presence of glutathione is known to derive thiyl radicals and to provide an additional important source of reactive oxygen species, thus propagating oxidative stress in MPO-rich cells (Borisenko et al., 2004). The reaction of thiyl radicals with GSH in the presence of oxygen leads to disulfide anion-radical formation, a reducing radical that can readily donate an electron to molecular oxygen to yield O2̇̄. Disproportionation of superoxide radicals leads to the accumulation of H2O2, a source of oxidizing equivalents for the MPO-catalyzed reactions. Hence, preferable oxidation of SH-groups by etoposide phenoxyl radicals may cause formation of oxygen radicals and etoposide redox-cycling implying the amplification of oxidative damage (and potentially recombinogenic oxidative DNA damage) in MPO-rich cells, including myeloid progenitor cells.
Rearrangements involving the MLL gene on chromosome band 11q23 are a hallmark of therapy-related acute myeloid leukemias after treatment with DNA topoisomerase II poisons including etoposide (Felix et al., 2006). Acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion (Libura et al., 2005). Etoposide promotes specific rearrangements of MLL in CD34+ consistent with the full spectrum of oncogenic events identified in leukemic samples. However, the mechanisms underlying etoposide-induced genotoxicity and leukemogenesis remain controversial. One possibility, as suggested above, is that MPO-catalyzed oxidation of etoposide leads to oxidative DNA damage, abasic sites, and resultant enhancement of DNA topoisomerase II-mediated MLL gene rearrangements. Others have suggested illegitimate recombination events in response to formation of DNA topoisomerase II covalent complexes may initiate MLL rearrangements (Sung et al., 2006). Any dysfunction or variation of DNA damage sensor and repair proteins might be expected to influence both the frequency and spectrum of repair products. In addition, when concentrations of endogenous GSH are low (either intrinsically or as a result of an oxidative process), MPO-catalyzed etoposide-O• formation may allow for direct oxidation of cysteines on topoisomerase II or other DNA-repair proteins that are essential for their functions. In particular, topoisomerase II is a strongly SH-dependent endonuclease. Its inhibition is associated with genotoxicity/mutagenicity and development of a procarcinogenic phenotype (Hutt and Kalf, 1996). Regardless of the mechanism(s) responsible for etoposide-induced leukemogenesis, our results demonstrate MPO dependence for both etoposide-induced DNA damage (Fig. 5) and for MLL gene rearrangements.
MPO is not the only enzyme that oxidizes etoposide. It has been suggested that oxidative activation of etoposide by cytochrome P450 monooxygenases, prostaglandin synthetase, and tyrosinase may contribute to its cytotoxicity (Haim et al., 1987; Usui and Sinha, 1990; Relling et al., 1994). However, these enzymes are not expressed appreciably in CD34+ cells (Fan et al., 2006). We hypothesize, therefore, that it is the oxidizing enzyme MPO in highly proliferative CD34+ cells that increases the risk of etoposide-induced secondary leukemias.
Oxidation of etoposide results in the formation of several metabolites (Fan et al., 2006; Zheng et al., 2006). Patients receiving etoposide accumulate appreciable levels of etoposide catechol catalyzed by the action of CYP3A4 and CYP3A5 (Zheng et al., 2004; Zhuo et al., 2004). Etoposide catechol can be converted to semiquinone radicals by one-electron oxidation catalyzed by MPO. MPO may also catalyze (in cells and in vitro) the formation of a two-electron oxidation product of etoposide, etoposide ortho-quinone that forms conjugates with GSH (Mans et al., 1992; Fan et al., 2006; Zheng et al., 2006) and may be responsible in part for the cyto- and genotoxic effects of etoposide because of its ability to stabilize topoisomerase II/DNA covalent complexes as studied in vitro (Lovett et al., 2001).
In conclusion, our study demonstrates MPO-dependent generation of etoposide-O• radicals in human myeloid progenitor CD34+ cells relevant to etoposide genotoxicity and carcinogenesis. EPR signals of etoposide phenoxyl radicals can be directly measured in these cells and can be used as a model to study MPO-induced oxidation of phenolic compounds. The data obtained should facilitate future studies of the mechanisms of etoposide-associated secondary leukemias and potentially lead to the development of nutritional antioxidant strategies to limit and/or prevent MPO-catalyzed formation of etoposide metabolites causative for therapy-related myeloid leukemias.
Authorship Contributions
Participated in research design: Vaslova, Feng, Gollin, Lewis, Kagan, and Yalowich.
Conducted experiments: Vaslova, Feng, Giorgianni, Do, Lewis, and Yalowich.
Contributed new reagents or analytic tools: Goff.
Performed data analysis: Vaslova, Feng, Giorgianni, Do, Gollin, Lewis, and Yalowich.
Wrote or contributed to the writing of the manuscript: Vaslova, Goff, Giorgianni, Do, Gollin, Lewis, Kagan, and Yalowich.
Acknowledgments
We thank Dr. Ching-Shih Chen and Dr. Mary K. Ritke for critical evaluation of this manuscript.
Footnotes
This research was supported in part by the National Institutes of Health National Cancer Institute [Grant R01-CA090787].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.068718.
-
ABBREVIATIONS:
- VP-16
- 4′-demethyl-epipodophyllotoxin-9-(4,6-O-ethylidene-β-d-gluco-pyranoside)
- MPO
- myeloperoxidase
- SA
- succinylacetone
- 3-AT
- 3-amino-1,2,4-triazole
- DTPA
- diethylentriaminepentaacetic acid
- CB
- human umbilical cord blood
- MLL
- mixed-lineage leukemia
- MLLR
- MLL gene rearrangements
- t-AML
- treatment-related acute myelogenous leukemia
- EPR
- electron paramagnetic resonance
- DMSO
- dimethyl sulfoxide
- PBS
- phosphate-buffered saline
- FBS
- fetal bovine serum
- FISH
- fluorescence in situ hybridization
- ACD-A
- anticoagulant citrate dextrose solution.
- Received September 7, 2010.
- Accepted November 19, 2010.
- Copyright © 2011 The American Society for Pharmacology and Experimental Therapeutics