Visual Overview
Abstract
G protein–coupled receptors (GPCRs), the largest family of signaling receptors in the human genome, are also the largest class of targets of approved drugs. Are the optimal GPCRs (in terms of efficacy and safety) currently targeted therapeutically? Especially given the large number (∼120) of orphan GPCRs (which lack known physiologic agonists), it is likely that previously unrecognized GPCRs, especially orphan receptors, regulate cell function and can be therapeutic targets. Knowledge is limited regarding the diversity and identity of GPCRs that are activated by endogenous ligands and that native cells express. Here, we review approaches to define GPCR expression in tissues and cells and results from studies using these approaches. We identify problems with the available data and suggest future ways to identify and validate the physiologic and therapeutic roles of previously unrecognized GPCRs. We propose that a particularly useful approach to identify functionally important GPCRs with therapeutic potential will be to focus on receptors that show selective increases in expression in diseased cells from patients and experimental animals.
Introduction
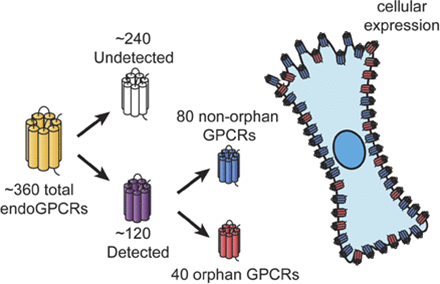
G protein–coupled receptors (GPCRs, also termed 7-transmembrane or heptahelical receptors) have been of major interest for investigators in many disciplines, including molecular pharmacology. Early studies on GPCRs assessed the action in cells and plasma membrane preparations of neurotransmitters, hormones, and pharmacological agents in terms of their ability to regulate the generation of second messengers (e.g., cAMP, Ca2+) and, in turn, cellular events via enzymes (e.g., protein kinases) and ion channels. Results obtained by the Human Genome Project and for the genomes of other eukaryotes have revealed that GPCRs are the largest family of signaling receptors in humans and other species (Fredriksson et al., 2003; Vassilatis et al., 2003; Insel et al., 2012; Foster et al., 2014b). The receptors include those that interact with endogenous ligands (endoGPCRs); GPCRs regulated by exogenous factors, such as photons of light, odorants, and tastants (chemosensory receptors); and GPCRs that lack known physiologic ligands (termed orphan receptors). It is estimated that among the approximately 800 GPCRs in humans, ∼380 are endoGPCRs, of which about one-third are orphan receptors, even though there have been substantial efforts at deorphanization (Fredriksson et al., 2003; Kroeze et al., 2003; Ozawa et al., 2010; Amisten et al., 2013; Civelli et al., 2013).
In parallel with work that has involved the cloning, genomic characterization, heterologous expression, and studies of GPCR actions and regulation, other efforts have emphasized the utility of GPCRs as therapeutic targets. Indeed, GPCRs are the largest class (∼30%) of the targets of approved drugs (Overington et al., 2006; Lundstrom, 2009; Rask-Andersen et al., 2014). Reasons for the utility of GPCRs as therapeutic targets include the many different types of chemical entities with which they interact, the accessibility of GPCRs on the plasma membrane from the extracellular milieu, their ability to initiate signaling pathways that undergo amplification in target cells, and the selectivity in their expression by different types of cells. This latter property aids in facilitating tissue- and cell-selective actions of GPCR-targeted drugs.
Despite the widespread use of GPCRs as therapeutic targets, one can ask the following: Are the optimal GPCRs (in terms of efficacy and safety) targeted by current therapeutic approaches? This question derives in part from the current therapeutic targeting of only a fraction of the endoGPCRs. Moreover, orphan GPCRs have largely been ignored as therapeutic targets. It is thus necessary to identify the repertoire of GPCRs—in particular, endoGPCRs—expressed by individual tissues and, more importantly, native cells. Studies to assess this gap in knowledge test the hypothesis that certain GPCRs are enriched in native cells, regulate cellular (and tissue) function, and can be targeted therapeutically. In this article, we review the approaches and data that have begun to provide information to test this hypothesis. In addition, we discuss problems and limitations of available data and future directions that may help definitively answer the question posed earlier.
Methods and Approaches to Assess GPCR Expression
Analyses of functional responses, second messengers, or other signaling events represent hypothesis-testing approaches by asking if a particular receptor is biologically active and provide indirect ways to assess GPCR expression by tissues and cells. Radioligand binding assays facilitate the direct identification and quantification of GPCRs. However, functional, signaling, and radioligand binding assays are biased approaches: one chooses a GPCR of interest and then uses agonists, antagonists, and radioligands for the receptor being assessed. Thus, one can only study receptors for which appropriate reagents are available.
By contrast, hypothesis-generating approaches are not based on prior knowledge of a GPCR being present, but instead rely on unbiased analyses of the expression of receptor mRNA or protein. Such approaches can define the GPCR expression profile/repertoire and can quantify receptor expression. Table 1 lists several approaches used to assess GPCR expression.
Unbiased methods to assess expression of GPCRs
Numerous studies have used DNA microarrays (“DNA chips”) to define the transcriptomes of cells and tissues. Such microarrays contain probes (specific DNA sequences) that hybridize with the genes of humans, mice, or other species. Hybridization of the probes to target genes is quantified by chemiluminescence, fluorescence, or another method, facilitating quantitation of the abundance of individual mRNAs/cDNAs. Commercially available microarrays that assess most or all genes in a transcriptome are not optimized to detect GPCRs, but such arrays have been used to characterize GPCR expression.
Proprietary and commercial GPCR microarrays, to be discussed later, and real-time polymerase chain reaction (PCR) analyses for individual GPCRs offer an alternative approach to identify chemosensory and nonchemosensory GPCRs. For example, Regard et al. (2008) used Taqman quantitative real-time PCR to quantify the transcripts of 353 nonodorant GPCRs in 41 adult mouse tissues and predicted previously unanticipated roles for less well studied receptors—an idea consistent with our hypothesis that previously unrecognized GPCRs are enriched in native cells, contribute to physiology, and are potential therapeutic targets.
With improvement in the ability to perform sequencing and a rapid decrease in its cost, new techniques such as high-resolution RNA sequencing have begun to be used to identify and quantify expression of GPCRs and other members of the transcriptome. Such studies have recently included the profiling of GPCRs expressed in single cells (Manteniotis et al., 2013; Spaethling et al., 2014).
An alternative to assessing the expression of GPCR mRNA is the use of unbiased proteomic approaches, such as mass spectrometry. Although such technology has not, as yet, been used to define and quantify overall GPCR expression in cells and tissues, initial results suggest this may be a feasible approach (Eisen et al., 2013; Feve et al., 2014).
Microarrays for the Detection of GPCRs
Commercial microarrays have been created that are optimized to detect and quantify the mRNA of ∼350 nonchemosensory GPCRs of mice, rats, and humans. We and others have found these arrays to be quite useful to assess GPCR expression. Figure 1 shows a comparison of the detection of GPCRs by such a targeted array (a Taqman GeneSignature array; Life Technologies, Carlsbad, CA) with results obtained using an Affymetrix Mouse Genome 430A microarray (Affymetrix, Santa Clara, CA) (which detects ∼14,000 mouse genes). We found that highly expressed GPCRs on the latter microarray show a positive correlation (and an R2 of 0.37) with the GPCR array data. However, assessment of GPCRs expressed at lower levels reveals that the Affymetrix array has many false-positive and false-negative results (and R2 = 0.01). GPCR arrays thus seem to be more useful to detect and quantify GPCRs than nontargeted arrays. Data from others support this conclusion (Maurel et al., 2011).
Comparison of GPCR expression data obtained using a GPCR-specific array and a generic cDNA array. The data were collected in two experiments, with data obtained using cDNA prepared from murine wild-type S49 lymphoma cells and that could be compared for GPCRs present on an Affymetrix Mouse Genome 430A microarray and a Taqman GeneSignature array. The 32 GPCRs highly expressed (>log26) on the Affymetrix array show a positive correlation (R2 = 0.37) between the two arrays. GPCRs expressed at lower levels (<log26) show numerous false-positive and false-negative results in the detection of GPCRs by the Affymetrix array compared with the GPCR array (R2 = 0.01). ΔCT, cycle threshold relative to the mRNA used to normalize expression of each GPCR; GCRMA, guanine cytosine robust multiarray analysis.
What Do Available Data Reveal about GPCR Expression in Tissues and Native Cells?
GPCR expression has thus far been determined in a number of tissues (Table 2) and a smaller number of individual cell types (Table 3). Our laboratory has assessed the expression of nonchemosensory GPCRs by numerous individual cell types, including lymphoid cells, dendritic cells, neutrophils, vascular smooth muscle cells, several types of fibroblasts, adipocytes, renal epithelial cells, trigeminal neurons, and several types of cancer cells.
GPCR expression in tissues
GPCR expression in cells
The notion that GPCRs and GPCR signaling pathways are altered (and are potential therapeutic targets) in cancer in addition to various endocrine tumors (for which GPCR-targeted drugs are commonly used) has been largely ignored by investigators in oncology. Even so, several recent reviews have emphasized the importance of GPCRs and GPCR signaling in cancer (Lappano and Maggiolini, 2011; Feigin, 2013; O'Hayre et al., 2013, 2014). O’Hayre and colleagues (2013) noted that nearly 20% of human tumors have GPCR mutations, 4% of tumors have activating mutations of the Gαs gene, and activating mutations of Gαq family members occur in melanomas. GPCRs may contribute to cancer not only by effects on the growth, death, metabolism, and function of malignant cells, but also by actions on cells of the tumor microenvironment, including vascular cells, immune cells, and cancer-associated fibroblasts. Although not widely explored, such actions may have functional importance for the malignant phenotype (Hanahan and Weinberg, 2011) and thus have therapeutic potential.
Several general conclusions regarding GPCR expression in tissues and cells can be drawn from published findings and our unpublished results:
Most tissues and individual cell types express at least 100 different GPCRs, including GPCRs that link to each of the major classes of heterotrimeric G proteins (Gs, Gi/o, Gq/11, and G12/13).
Among the GPCRs with the highest expression are those not previously known to be expressed in the previously mentioned tissues and cells and, thus, not the subject of prior studies.
Many of the highest expressed GPCRs are orphan receptors.
Cells from animals or patients with particular diseases show prominent changes (increases and decreases) in expression of numerous GPCRs, thus revealing disease-specific changes in GPCR expression.
Additional methods [quantitative real-time PCR, antibody-based and functional (including signal transduction and cellular response) assays]) confirm data obtained with GPCR arrays, including, for example, evidence that the expression of several Gs-coupled GPCRs correlates with their ability to maximally stimulate cellular cAMP production.
Such findings thus indicate that analysis of GPCR expression of tissues, and especially of individual cell types, appears to be a highly useful means to identify previously unrecognized GPCRs that may be functional, contribute to pathophysiology, and serve as novel drug targets.
What Are Some of the Problems and Limitations of Efforts to Define GPCR Expression in Tissues and Cells?
Despite the potential importance of the findings related to the discovery of “new GPCRs,” i.e., endoGPCRs not previously known to be expressed, in individual tissues and cell types, a number of issues must be considered in studies of GPCR expression:
The source of material analyzed for GPCR expression: The results in Table 2 were primarily derived from studies of whole organs, tissues, and tissue biopsies, all of which are heterogeneous cellular preparations. The contribution of the different cell types in such preparations is thus not clear. Studies of individual cell types (Table 3) obviate this concern. Even in such studies, however, one analyzes a population of cells. The use of techniques such as high-resolution RNA sequencing (Manteniotis et al., 2013; Spaethling et al., 2014) and perhaps proteomic methods (Davies et al., 2007; Wu et al., 2012; Eisen et al., 2013) to assess GPCRs in individual cells will thus be an important future direction for defining the profile and variability in GPCR expression in particular cell types.
Reproducibility and consistency of methods used to prepare mRNA, cDNA, or protein and to assess gene and protein expression: Standards have been developed to facilitate reproducibility in microarray (Brazma et al., 2001; Hansen et al., 2007; Rung and Brazma, 2013) and proteomic studies (Davies et al., 2007; Taylor et al., 2007), but results from different laboratories can differ. Such differences are generally attributed to variations in the methods used for the preparation and processing of samples. Primer design may not be ideal for the detection of all relevant GPCRs by arrays. For example, Taqman-based GPCR arrays may not detect certain, including functionally relevant, GPCRs. The use of different approaches for data analysis and statistics by different laboratories can contribute to discordant results (Pan, 2002; Motulsky, 2014). The dearth of well validated antibodies to detect native GPCRs, including for use in proteomic studies, is an important problem in GPCR research (Hutchings et al., 2010; Talmont et al., 2012; Eisen et al., 2013; Marchalant et al., 2014). Use of antibodies to study GPCRs is challenging because the receptors are typically expressed in cells at lower levels than many other cellular proteins. Criteria, including studies with cells or animals engineered to have a knockout of a particular GPCR, have been proposed to help validate GPCR-targeted antibodies (Michel et al., 2009). Moreover, since protein levels in cells are not necessarily predicted by mRNA abundance, differences in protein expression could derive from factors that include altered protein translation and/or degradation (de Sousa Abreu et al., 2009; Maier et al., 2009). Especially given the difficulties in obtaining well validated GPCR antibodies, a possible solution is to measure levels of actively translated mRNA using polysome profile analysis together with DNA high-density arrays or GPCR-specific quantitative PCR (Mašek et al., 2011; Gandin et al., 2014).
Normalization of results for GPCR expression: Most assays used to determine mRNA and protein expression rely on normalization to “housekeeping” genes/proteins. An extensive literature has discussed problems related to normalization in such studies. One such problem is a change in expression of the gene/protein used for normalization—a particular concern in studies of development, differentiation, or disease (Khimani et al., 2005; Brattelid et al., 2007). Two issues in the use of arrays to assess GPCR expression are how to define the limit of detection of a receptor in terms of ∆cycle-threshold [∆C(t)] relative to the reference used for normalization, and the most appropriate statistical tests for data analysis (Khimani et al., 2005; Rubie et al., 2005).
Approaches used to classify GPCRs in profiles of their expression in tissues and cells: One approach is to cluster results for GPCR expression on the basis of the coupling of receptors to heterotrimeric G proteins. Such information is available in articles and databases, such as the International Union of Basic and Clinical Pharmacology/British Pharmacological Society Guide to Pharmacology (Pawson et al., 2014). The International Union of Basic and Clinical Pharmacology/British Pharmacological Society systematically annotates each GPCR and provides reviews from expert subcommittees for each of the target families in the database. Results that supersede such data are published on an ongoing basis; therefore, one must be vigilant in updating conclusions regarding the coupling of GPCRs to G proteins. Problems associated with classifying results on this basis include the evidence that some GPCRs couple to multiple G proteins, the limited data regarding the G protein linkage of orphan GPCRs, and the ability of GPCRs to act via β-arrestin instead of (or in addition to) G proteins. Other ways to classify GPCRs include the Glutamate, Rhodopsin, Adhesion, Frizzled/taste2, and Secretin system, evolutionary relationships, ligand interactions, structural data, and susceptibility to post-translational modifications (Fredriksson et al., 2003; Davies et al., 2007; Secker et al., 2010; Lin et al., 2013; Venkatakrishnan et al., 2013).
Which types of GPCRs should be studied? As shown by the studies in Tables 2 and 3, most efforts have focused on the use of array- or PCR-based methods and have emphasized the expression of nonchemosensory GPCRs. Recent studies, however, have identified chemosensory receptor expression in tissues not typically thought to be in involved in sensation (Reimann et al., 2012; Foster et al., 2014a,b; Pronin et al., 2014; Rajkumar et al., 2014; Malki et al., 2015). With the discovery of small-molecule metabolites [in some cases, products of microbiota (e.g., Natarajan and Pluznick, 2014)] that interact with GPCRs and that may have been considered odorants or tastants, the distinction between chemosensory and nonchemosensory GPCRs is blurring. Thus, future studies may need to incorporate analyses of both types of GPCRs.
Prioritization of results regarding GPCR expression to guide subsequent studies that validate the expression and evaluate the functional role of individual receptors: The discovery that native cells typically express >100 GPCRs creates an embarrassment of riches, but also a challenge in terms of choosing individual GPCRs for subsequent studies. A key goal is to identify GPCRs that are important for cellular function and that may be therapeutic targets. The use of RNA interference and gene editing approaches (i.e., CRISPR/Cas9) to knock down receptor expression in cells of interest and then to assess the impact of receptor knock down on functional activity (signal transduction or cellular responses) provides a way to survey a population of GPCRs and identify physiologically (and potentially pharmacologically) important GPCRs (Willets and Nash, 2013). Additional features to help such prioritization include 1) the level of expression (more highly expressed GPCRs are predicted to contribute to a greater extent to cell function and may be easier to study); 2) knowledge of other cell types that express receptors of potential interest [to achieve greater selectivity in the site of action, including by comparing expression in cells that are related to one another, such as, for example, in different types of macrophages (Lattin et al., 2008; Groot-Kormelink et al., 2012; Hohenhaus et al., 2013)]; 3) choosing GPCRs whose expression is shared in a cell type found in humans and experimental animals (e.g., mice, rats); and 4) availability of reagents (e.g., agonists, antagonists, antibodies) to conduct signaling and functional studies. A useful feature, especially for studies of disease settings, is to focus efforts on GPCRs with prominent differences in expression between the normal cells and cells from diseased animals or patients. Differential GPCR expression can also be helpful in choosing GPCRs for studies of development and differentiation of tissues and cells.
Determining the function of highly expressed orphan GPCRs: Some of the most exciting (and unexpected) data that we have obtained from our studies of GPCR expression in native cells is the high expression of a variety of orphan GPCRs. This is perhaps not surprising since orphan receptors represent about one-third of the endoGPCRs, and limited reagents have been available to assess function mediated by most orphan GPCRs. Since the endogenous ligands for such receptors are unknown, investigating their physiologic roles can be difficult (Ahmad et al., 2014). Molecular approaches, such as RNA interference or overexpression in heterologous cells, can help define signaling mechanisms, especially if a GPCR is constitutively active. Antibodies to orphan GPCRs might be used to block signaling and obviate the need for a ligand, although, as noted earlier, validation of GPCR antibodies can be difficult. Another challenge with orphan GPCRs has been the difficulty in setting up screens for ligands, as the G protein coupling of an orphan may not be known; chimeric G proteins may be a way to develop high-throughput screens (Yin et al., 2004).
Summary and Conclusions
Even though they have been widely studied and highly useful as therapeutic targets, GPCRs continue to be very important molecular entities. The recognition from genomic studies that there are many more GPCRs than were previously known or characterized, and that many of these are orphan receptors provides opportunities to discover physiologic and therapeutic roles for newly recognized GPCRs. Studies of GPCR expression in tissues, but especially in native cells, can reveal that previously unrecognized GPCRs contribute to cell function in health and disease. Numerous challenges exist in studies of GPCR expression, but if “new” GPCRs can be validated and shown to be functionally active, we anticipate that such GPCRs may prove to be as important—and perhaps even more so—than the GPCRs that have been the focus of efforts in physiology and pharmacology, and that are so valuable as therapeutic targets.
Acknowledgments
The authors thank their collaborators in recent efforts of their laboratory related to GPCR expression: Alessandra Franco, Randall French, Thomas Kipps, Jihyung Lee, Andrew Lowy, Victor Nizet, Hemal Patel, Laura Rassenti, Eyal Raz, Patricia Thistlethwaite (all from the University of California, San Diego), Christopher Walker, Debbie Hay (University of Auckland, Auckland, New Zealand), Michael Feigin (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY), Ana Kilic, and Alexander Pfeifer (University of Bonn, Bonn, Germany).
Authorship Contributions
Conducted experiments: Wilderman, Zambon.
Performed data analysis: Wilderman, Zambon.
Wrote or contributed to the writing of the manuscript: Insel, Wilderman, Zambon, Snead, Murray, Aroonsakool, McDonald, Zhou, McCann, Zhang, Sriram, Chinn, Michkov, Lynch, Overland, Corriden.
Footnotes
- Received January 23, 2015.
- Accepted March 2, 2015.
Work in the authors’ laboratory on this topic has been supported by research and training grants from the National Institutes of Health [Grants CA189477, CA121938, GM68524, HL091061, HL066941, HL007444, and GM-68524], the Department of Defense [Grant W81XWH-14-1-0372], and with financial assistance from Roche, Pfizer-CovX, Bristol Myers Squibb, the American Heart Association, and an ASPET-Astellas Award in Translational Pharmacology.
Abbreviations
- endoGPCR
- GPCR activated by endogenous ligands
- GPCR
- G protein–coupled receptor
- PCR
- polymerase chain reaction
- Copyright © 2015 by The American Society for Pharmacology and Experimental Therapeutics