Abstract
Human breast cell carcinoma MCF-7 cells were found to bind125I-labeled rat amylin (rAmylin) and the peptide amylin antagonist radioligand 125I-AC512 with high affinity. This high affinity binding possessed characteristics unique to the already defined high affinity binding site for amylin in the rat nucleus accumbens [Mol. Pharmacol. 44:493–497 (1993);J. Pharmacol. Exp. Ther. 270:779–787 (1994);Eur. J. Pharmacol. 262:133–141 (1994)]. To further define this receptor, we report results of expression cloning studies from an MCF-7 cell library. We isolated two variants of a seven-transmembrane receptor that were identical to two previously described human calcitonin receptors (hCTR1 and hCTR2). These receptors were characterized by expression in different surrogate host cell systems. Transient expression of hCTR1 in COS cells yielded membranes that bound 125I-AC512 and 125I-salmon calcitonin with high affinity, but no high affinity binding was observed with 125I-human calcitonin (hCAL) or125I-rAmylin. Stable expression of hCTR1 in HEK 293 cells produced similar data. In contrast, expression of hCTR2 in COS cells yielded membranes that bound 125I-AC512,125I-hCAL, and 125I-rAmylin with high affinity. The agonists 125I-hCAL and 125I-rAmylin bound 65% and 1.5%, respectively, of the sites bound by the antagonist radioligand 125I-AC512 in this expression system. This pattern of binding was repeated in HEK 293 cells stably transfected with hCTR2 (125I-hCAL = 24.8%B max, 125I-rAmylin = 8%B max). In both expression systems, the agonists hCAL and rAmylin were much more potent in displacing their radioligand counterparts than was the antagonist radioligand125I-AC512. For example, the pK i value for displacement of125I-AC512 by rAmylin was 7.2 in HEK 293 cells but rose to 9.1 when displacing 125I-rAmylin. Finally, hCTR2 was expressed in baculovirus-infected Ti ni cells. In this system, only specific binding to the antagonist 125I-AC512 and agonist 125I-hCAL was observed; no binding to125I-rAmylin could be detected. These data are discussed in terms of two working hypotheses. The first is that amylin is a weak agonist for hCTR2 and that this receptor is unrelated to the amylin receptor found in this cell line. The second is that hCTR2 couples to different G proteins for calcitonin and amylin function in different cells. At present, these data cannot be used to disprove conclusively either hypothesis.
Amylin is a peptide hormone that is synthesized and secreted from pancreatic β cells. There is evidence that an increase in the cosecretion ratio of amylin to insulin exacerbates insulin tolerance, and in general, there are data to suggest that this peptide may be important in the pathology of type II diabetes (1-5). High affinity binding for125I-rAmylin has been reported in the rat nucleus accumbens (6-8), thus defining an operational and experimentally accessible amylin receptor. Similar high affinity binding of both125I-rAmylin and an sCAL antagonist analogue radiolabel 125I-AC512 has been described (9) in human MCF-7 cells. These data raise the possibility that these cells contain a human amylin receptor, and this information would be of value in the study of the role of amylin in human type II diabetes.
We describe the expression cloning of the125I-AC512 binding site from an MCF-7 cell cDNA library and the subsequent identification of the gene products as previously classified CTRs (10). The receptor pharmacology of these gene products in various host cells is described, as are data to suggest a relationship between the hCTR and the responses of human systems to amylin.
Materials and Methods
Cell culture.
MCF-7 human breast adenocarcinoma cells from pleural effusion (HTB 22; American Type Culture Collection, Rockville, MD) were cultured in Eagle’s minimal essential medium with nonessential amino acids, sodium pyruvate [1 mm (90%)], and fetal bovine serum (10%). Cells were grown as a monolayer, fed fresh media every 3 days, and split 1:2 weekly. Transfected COS-7 or HEK 293 cells were also cultured in Eagle’s minimal essential medium with nonessential amino acids, sodium pyruvate [1 mm(90%)], and fetal bovine serum (10%). The cells were grown as a monolayer, fed fresh media every 3 days, and split 1:2 weekly.
Membrane preparation.
The nucleus accumbens was dissected from the brains of Sprague-Dawley rats (200–250 g) and homogenized (three 15-second bursts) in ice-cold HEPES buffer (20 mmHEPES, pH adjusted to 7.4 with NaOH at 23°). The homogenate was centrifuged at 48,000 × g for 15 min and washed twice by resuspension in fresh buffer. The membrane pellet from the third centrifugation was resuspended in fresh buffer with 0.2 mmPMSF, aliquoted, and stored at −70°. The MCF-7 cells were harvested at confluency by manual scraping of the tissue culture flasks. The cells then were pelleted by centrifugation at 2000 rpm for 15 min and homogenized as described above.
Cultured cells were harvested at confluency by manual scraping of the tissue culture flasks, then pelleted by centrifugation at 2000 rpm for 15 min, and homogenized (three 15-second bursts) in ice-cold HEPES buffer (20 mm HEPES, pH adjusted to 7.4 with NaOH at 23°). The homogenate was centrifuged at 48,000 × gfor 15 min and washed twice through resuspension in fresh buffer. The membrane pellet from the third centrifugation was resuspended in fresh buffer with 0.2 mm PMSF, aliquoted, and stored at −70°.
Synthesis of AC512.
To a solution of the peptide (Ac)LGKLSQELHRLQTY-PRTNTGSNTY(NH2) (128.5 mg, 80% peptide content, 36.4 μmol; synthesized using normal solid-phase techniques from Fmoc-protected amino acids and a Rink resin) in 25 mm aqueous NaHCO3 (30 ml, pH ∼ 8) maintained at 0° we added dropwise a solution of 14.2 mg of cold Bolton-Hunter reagent [3-(3-iodo-4-hydroxyphenyl)-propanoic acidN-hydroxysuccinimide ester] in 8 ml of acetonitrile. The resulting solution was stirred for 2 hr at 0°. An additional 10 mg of the Bolton-Hunter reagent in 5 ml of acetonitrile was added at this point, and the solution was stirred for 1 additional hr at 0°. The reaction was quenched by the addition of sufficient 10% aqueous trifluoroacetic acid to give pH 1.5 and warmed to room temperature. This solution was filtered to remove a small amount of precipitated material. The filtrate was injected onto a Waters (Milford, MA) Delta-Prep HPLC equipped with a radial compression C-18 cartridge (conditions: flow rate, 100 ml/min; solvent A = CH3CN, solvent B = 0.1% aqueous trifluoroacetic acid; initial conditions were 73% B; 4 min after the injection a linear gradient was begun, decreasing the percentage of B to 53% over 40 min). The desired product, AC512, eluted at 22.5 min. Lyophilization gave 89.2 mg of AC512 as a white powder. Peptide content was found to be 64.5%. High resolution mass spectrum (electrospray): expected monoisotopic MH+ 3092.4091, found: 3092.4423. The most likely spot for derivatization of the starting peptide is on the lysine, and AC512 is assigned the structure (Ac)LG(KBH)LSQELHRLQTYPRTNTGSNTY(NH2) with (KBH) being lysine labeled on the ε-amino group with the Bolton-Hunter reagent. The 125I-labeled material was synthesized at Amersham (Arlington Heights, IL) using the same starting peptide and radioactive Bolton-Hunter reagent. The Amersham125I-labeled peptide coeluted with the nonradioactive peptide synthesized as described above.
Receptor binding.
Membranes were incubated with125I-rAmylin (Bolton Hunter labeled at the amino-terminal lysine; Amersham) or 125I-AC512 (Bolton Hunter-labeled [Arg18,Asn30,Tyr32]9–32 sCAL, 2000 Ci/mmol; Amersham) in 20 mm HEPES buffer, containing 0.5 mg/ml bacitracin, 0.5 mg/ml bovine serum albumin, and 0.2 mm PMSF (all from Sigma Chemical, St. Louis, MO) plus test ligand or ligands, for 60 min at 23° (samples mixed on a Titer Plate Shaker; Lab-Line Instruments). Nonspecific binding was defined as the radioactivity remaining in the presence of 100 nm sCAL. Incubations were carried out in triplicate tubes and were started by the addition of membrane. Binding was terminated by filtration through glass-fiber filters (presoaked 30 min in 0.5% polyethyleneimine), using the Skatron semiautomatic cell harvester. Filters were placed in Sarsted 68.752 51 × 12 mm polypropylene tubes and counted for 1 min in a γ-counter.
Binding data were analyzed with the Glaxo Wellcome statistical fitting package RADLIG (Glaxo Wellcome Scientific Computing) to simultaneous equations describing total binding (saturable binding according to a logistic equation plus a linear nonspecific binding curve) and a linear nonspecific binding curve. Statistical analyses were used to determine whether data could be fit to a single or double population of binding sites. Saturation analysis yielded a nonlinear least-squares fit to the logistic equation with a half-maximal fitting parameter (the equilibrium dissociation constant of the ligand/receptor complex under ideal conditions, denoted Kd ) and a maximal asymptote (denoted B max, providing an estimate of the maximal number of binding sites in fmol/mg of protein). Displacement analysis (radioligand concentrations = 0.3 × Kd ; incubations for 90 min) yielded concentrations of nonradioactive ligand that half-maximally displaced a given concentration of radioligand (denoted IC50). This value was used to calculate an estimate of the equilibrium dissociation constant of the nonradioactive ligand/receptor complex (denoted Ki ) by correction for the amount of radioactive ligand and theKd value (11). Complex displacement curves were fit to a two-population model, yielding two apparent affinities and the relative quantities of two apparent sites (or receptor states).
Molecular biology.
Standard molecular biology techniques were used (12). Poly(A)+ RNA was isolated using a FastTrack RNA isolation kit (InVitrogen, San Diego, CA). Both strands of the CTR cDNA were sequenced with an ABI394 automatic sequencer with use of the Analysis (Applied Biosystems, Foster City, CA) and Assembly LIGN (Kodak IBI, New Haven, CT) software programs.
Construction of cDNA library.
Five micrograms of poly(A)+ RNA, isolated from MCF-7 cells, was used to construct a size-selected cDNA library according to the manufacturer’s protocol (InVitrogen). Double-stranded, oligo(dT)-primed cDNA was synthesized and ligated toNotI/EcoRI adaptors. Fragments of cDNA of >1.6 kb were isolated and subsequently ligated into the EcoRI site of expression vector pMT4 (13). Aliquots of the library were titered by electroporation into Top10 competent cells (Stratagene, La Jolla, CA). Pools of colonies, representing ≈1000 independent cDNAs/pool, were scraped from plates and grown for 4–6 hr in 25 ml of LB-ampicillin. Plasmid DNA from each pool was isolated using the Wizard Midiprep DNA purification system (Promega, Madison, WI).
Expression cloning.
Pools of cDNA were transfected into COS-7 cells and analyzed for their ability to bind125I-sCAL or 125I-AC512 in transient transfection assays. Initial experiments were done with125I-sCAL; however, in view of the nearly irreversible kinetics of this radioligand, later experiments were done with 125I-AC512. On day 0, 3–4 × 105 COS-7 cells/well were plated onto six-well dishes. On day 1, cells were transfected with 1 μg of DNA from each pool per well using lipofectamine (GIBCO BRL, Gaithersburg, MD) according to the manufacturer’s instructions. Cells were assayed 48 hr after transfection by binding analysis (see below). After screening 0.6 × 106 clones, four pools that bound125I-ligand were identified. One pool was progressively subdivided into smaller pools until a single positive clone was obtained.
Generation of stable cell lines.
On day 0, HEK 293 cells were plated at a concentration of 106cells/100-mm dish. On day 1, cells were cotransfected with clone 77/pMT4 or clone 134/pMTR with pRSV/neo at a 10:1 ratio, respectively, according to the calcium phosphate method (Promega). On day 3, transfected cells were selected using G418-supplemented media at a concentration of 600 μg/ml. After a 2-week selection period, several colonies from each transfection were selected and expanded. Stable lines were checked for expression by binding of125I-sCAL or 125I-AC512.
Baculovirus Ti ni cells.
TheBglII/NotI fragment of CTR cDNA was subcloned into pFASTBACI vector (GIBCO BRL). Recombinant baculovirus was generated according to Life Technologies Bac-To-Bac Baculovirus Expression System Manual (14) efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediatedExpression System Manual. Spodoptera frugiperda(Sf9) cells (American Type Culture Collection) were used for transfection, virus amplification, and titering and were grown in supplemented Grace’s insect culture medium (GIBCO BRL) with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 1% pluronic F-68 (GIBCO BRL), and 50 mg/ml gentamycin (GIBCO BRL). Ti nicells (gift from JRH Biosciences, Lenexa, KS) were used for recombinant protein generation and were grown in Ex cell 405 insect medium (JRH Biosciences) with 50 mg/ml gentamycin. Using the initial transfection mix harvested 48 hr after infection, recombinant viruses were amplified in Sf9 cells and titered. Ti ni cells at 1.2 × 106 cells/ml were infected at a multiplicity of infection of 2 plaque-forming units/cell and were harvested 48 hr after infection. Cells pellets were washed once with phosphate-buffered saline and then frozen until assayed.
Microphysiometry.
This technique is based on the principle that the metabolism of cells is tightly linked to hydrogen ion output. A thin disk of cells is cultured over a pH detector and perfused with medium. Although perfusion takes place, the pH registered by the detector is constant. At regular intervals, the perfusion is stopped and the hydrogen ion allowed to accumulate in the chamber. The resulting decrease in pH with time is measured as a rate; this rate is proportional to the metabolic state of the cell. The overall cellular metabolism is measured as a succession of rates of secretion of hydrogen ion.
At ≈16 hr before the experiment, cells were seeded (300,000 cells/chamber) at 75–85% confluency in microphysiometer capsule cups. Capsules were then kept in a CO2 incubator at 37°. For experimental procedures, the microphysiometer was primed with low buffer media (modified RPMI 1640 medium; Molecular Devices, Menlo Park, CA) for 10 min, sensor chambers were put in place, and cell capsules were placed in the sensor chambers. After calibration of the microphysiometer, cells were allowed to equilibrate in a constant flow of media for 30–60 min to attain a steady base-line of hydrogen ion output. Perfusion was then changed to media containing the test drugs and effects recorded by computer .
Results
Rat nucleus accumbens binding.
High affinity reversible binding was observed with the radiolabel sCAL analogue125I-AC512 and125I-rAmylin. Binding reached steady state by 60–120 min and was displaceable with nonradioactive sCAL and amylin.125I-AC512 showed considerably less filter binding and nonspecific binding than125I-rAmylin. Over a range of concentrations,125I-AC512 yielded much higher specific binding in the rat nucleus accumbens than 125I-rAmylin. In addition to these improved binding characteristics and in contrast to 125I-rAmylin, 125I-AC512 is an antagonist with no observable agonist activity in amylin functional tissue systems, thereby reducing the possible complication of binding effects by G protein coupling.
125I-AC512 bound with high affinity to membranes prepared from rat nucleus accumbens in a saturable manner (see Table1for saturation binding data). Bound 125I-rAmylin could be displaced with agonists and antagonists. Table2 shows the chemical structures of the antagonists (AC512, AC66, AC413, hCGRP8–37) and agonists (rAmylin; human, rat, human, and eel CAL; and rCGRP). The equilibrium dissociation constants of the nonradioactive displacing ligand/receptor complex (Ki ) for the displacement of 125I-amylin are given as negative log values (pKi ) (see Table3). In general, all displacement curves were monophasic with Hill coefficients not significantly different from unity.
Rat nucleus accumbens: saturation binding
Peptide antagonists and agonists
Rat nucleus accumbens: displacement of [125I]amylin
Bound 125I-AC512 also could be displaced with nonradioactive ligands. The data describing the displacement by these ligands are given in Table4. In contrast to the simple curves obtained with the antagonists, displacement of 125I-AC512 with agonists produced complex biphasic curves. The data could be fit with a two-population model; the apparent affinities for the two populations and their relative abundance are given in Table 4. As seen from these data, the affinity of the agonists varied with the radioligand used. When125I-rAmylin was displaced, the agonists produced monophasic curves with high affinities. In contrast, during displacement of 125I-AC512, biphasic curves with two apparent affinities were obtained.
Rat nucleus accumbens: displacement of [125I]AC512
Human MCF-7 cells.
The human breast carcinoma cell line MCF-7 binds 125I-rAmylin with high affinity; this was confirmed in the current study with membrane from MCF-7 cells. The quantitative data describing this binding activity are given in Table5. 125I-rAmylin bound with affinity similar to that found in the rat nucleus accumbens, but the estimate for the maximal number of 125I-rAmylin binding sites in MCF7–7 cells was considerably larger (155.2 fmol/mg of protein; 95% CI, 64–246 fmol/mg of protein). As was seen in the rat nucleus accumbens, 125I-AC512 bound with high affinity (77.6 pm; 95% CI, 50–123 pm) and a greaterB max value (337 fmol/mg of protein; 95% CI, 180–493 fmol/mg of protein) than that found for125I-rAmylin.
MCF-7 cells: saturation binding
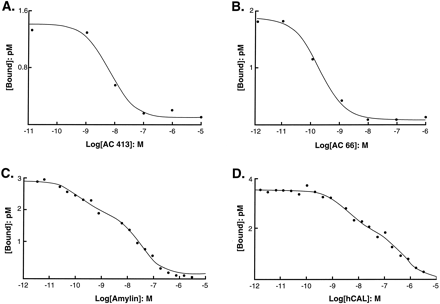
A range of peptide antagonists and agonists displaced125I-rAmylin from the binding sites, all with monophasic displacement curves with Hill coefficients not significantly different from unity (Fig. 1, A and B). Table 6shows pKi estimates from displacement studies. The affinity of AC512, AC66, and AC413 was comparable to that found for the rat nucleus accumbens. Of note was the 30-fold loss in potency in the MCF-7 cell over the rat nucleus accumbens for hCGRP8–37.
Displacement of bound 125I-AC512 with nonradioactive antagonist (A and B) and agonist (C and D) ligands in MCF-7 cells. [Bound], bound radioactivity.Log[Amylin], logarithms of molar concentrations of nonradioactive ligand. Displacement by AC413 (A), AC66 (B), rAmylin (C), and hCAL (D).
MCF-7 cells: displacement of [125I]amylin
As with rat nucleus accumbens, differences in potencies of displacing ligands were observed with the displacement of bound125I-AC512. Although monophasic single displacement curves were obtained for the antagonists, complex displacement curves were obtained for agonists (Fig. 1, Cand D). These curves yielded two apparent affinities and two apparent binding site populations (Table 7).
MCF-7 cells: displacement of [125I]AC512
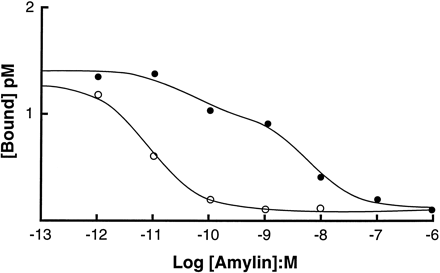
The pKi values in membranes from MCF-7 cells for agonists differed for displacement of125I-rAmylin and125I-AC512, as observed in the rat nucleus accumbens. A composite experiment of the differences in the displacement curves with rAmylin against125I-rAmylin and 125I-AC512 is shown in Fig. 2. The levels of initial radioligand binding were comparable, but the location parameters for the displacement curves differed by a factor of nearly 100.
Comparison of displacement of125I-rAmylin (□) and 125I-AC512 (•) with nonradioactive rAmylin in MCF-7 membranes. [Bound], bound radioactivity. Log[Amylin], logarithms of molar concentrations of nonradioactive ligand.
Expression cloning.
Transiently transfected COS-7 cells cultured in six-well plates were exposed to125I-sCAL or 125I-AC512 (≈20,000 cpm/well). Both radiolabels were used in separate experiments to maximize the possibility of obtaining a specific amylin-binding protein. A total of 171 pools screened yielded 4 pools exhibiting 125I-sCAL and125I-AC512 binding. From positive pool 77, an additional 20 pools of ≈100 clones each were prepared, and 2 subpools were found to be positive on further assay. The positive subpool was subfractionated until a single clone (clone 77) was isolated. A search of the sequence database revealed that the DNA and the deduced amino acid sequence for clone 77 were identical to those for hCTR1 (15). The only difference between clone 77 and the published sequences was a 78-bp segment missing at the 5′ untranslated region of clone 77. Insertion of the 78-bp segment created an in-frame ATG by the addition of 26 extra amino acids at the amino terminus of the CTR.
Using the polymerase chain reaction with CTR-specific primers, the three remaining pools were examined and determined to contain the CTR cDNA. Clone 77 cDNA then was used to screen the three positive pools by colony hybridization to isolate additional clones. Sequences of these three clones (clones 40, 134, and 167) revealed different lengths for the 5′- and 3′-untranslated regions but with no apparent difference in the coding region. Deduced protein sequences were identical to those of hCTR2 isolated from T47D cells (10). Clone 134 was the longest cDNA among the three and was chosen for further characterization. Comparison of clones 77 and 134 revealed that clone 77 contained a 16-amino acid insert in the first intracellular loop that was absent in clone 134. The differences between the two cDNAs are shown in Fig.3.
cDNA for clones 77 and 134 shows the similarity to the CTR cloned by Gorn et al.. (15) from BIN-67 cells.
Receptor binding: clone 77 (hCTR1).
hCTR1 transfected into COS-7 cells produced saturable binding with125I-AC512 and 125I-sCAL. For 125I-AC512, the equilibrium dissociation constant (Kd ) was 71 pm (95% CI, 30–158 pm) with a B max estimate of 597 ± 66 fmol/mg of protein (95% CI, 470–722 fmol/mg of protein; four experiments). This maximal number of binding sites was confirmed with saturation binding with 125I-sCAL (680 fmol/mg of protein; 95% CI, 540–816; three experiments). TheKd value for125I-sCAL was 2.8 pm (95% CI, 0.7–10.5 pm). Displacement experiments indicated that 125I-AC512 could be displaced by sCAL (pKi = 11.16), AC512 (pKi = 10.4), and hCGRP8–37 (pKi = 10.4). In view of these data, clone 77 was transfected into HEK 293 cells, and a stable cell line expressing hCTR1 was made.
As shown in Table 8, hCTR1 expressed in HEK 293 cells furnished membranes that saturably bound 125I-AC512 with aKd value of 200 pm (95% CI, 125–316 pm) and a B max value of 1493 ± 276 fmol/mg of protein (95% CI, 970-2017). These membranes also saturably bound 125I-sCAL with aKd value of 3.5 pm (95% CI, 2.7–4.6 pm). The B max value of 1260 ± 360 fmol/mg of protein (95% CI, 576-1944) was not significantly different from that found for the binding of 125I-AC512. No appreciable binding of 125I-hCAL or125I-rAmylin was observed. The potency of nonradioactive antagonists and agonists in displacing125I-AC512 is shown in Table 9. No high affinity binding was observed with agonists in these membranes.
HEK 293 cells (clone 77): saturation binding
HEK 293 cells (clone 77): displacement of [125I]AC512
Receptor binding clone 134 (hCTR2): COS-7 cell membranes.
125I-AC512 bound with high affinity to membranes prepared from COS-7 cells transiently transfected with hCTR2 cDNA. The equilibrium dissociation constant of the AC512/receptor complex was 245 pm (95% CI, 165–370 pm; see Table10). The maximal number of binding sites was 4423 fmol/mg of protein (95% CI, 3603–5242). Binding of somewhat higher affinity was observed with125I-sCAL. TheKd value was 28.2 pm (95% CI, 7–110 pm). The B max value was 5225 fmol/mg of protein (95% CI, 4235–6214), a value not significantly different from that found for 125I-AC512 (Table 10).
COS-7 cells (clone 134): saturation binding
The difference between hCTR1 and hCTR2 was that the latter, when transiently transfected into COS-7 cells, also bound125I-hCAL in a saturable manner (Kd = 417 pm;95% CI, 269–655 pm). In contrast to the data with 125I-AC512 and125I-sCAL, the B maxvalue for 125I-hCAL binding was significantly lower (3025 fmol/mg of protein; 95% CI, 1898–4150).
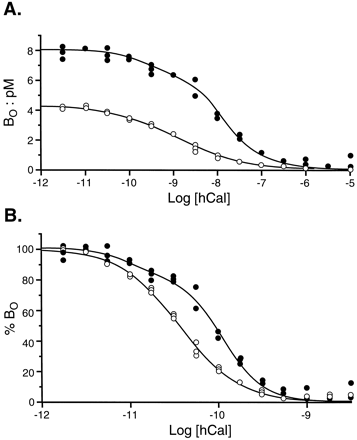
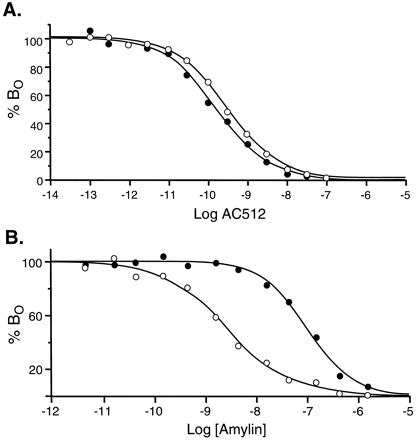
Bound 125I-AC512 and125I-hCAL could be displaced with agonists and antagonists for CTRs. Although the pKi estimates for the displacement of both radioligands with antagonists were uniform (Table 11), hCAL had a higher estimated affinity for the displacement of125I-hCAL (as opposed to125I-AC512). Fig. 4shows the displacement of 125I-AC512 and125I-hCAL by hCAL. It can be seen from this figure that the curve for displacement of125I-hCAL is monophasic, whereas that for displacement of 125I-AC512 is biphasic and shifted to the right.
COS-7 cells (clone 134): displacement of radiolabels
Displacement of 125I-AC512 (•) and125I-hCAL (○) by hCAL in membranes from COS cells. A,[B O ], bound radioactivity. Log[hCal], logarithms of molar concentrations of nonradioactive ligand. B, Data in A with ordinate values recalculated as a percentage of the initial BObinding of radioligand.
Membranes from COS-7 cells transiently transfected with cDNA for hCTR2 also saturably bound 125I-rAmylin (Kd = 275 pm; 95% CI, 30–2000 pm). TheB max value for125I-rAmylin binding was much lower (65 fmol/mg of protein; 95% CI, 36–94) than that found for125I-AC512, 125I-hCAL, or125I-sCAL. A limited displacement study in COS-7 cell membranes indicated that the saturable125I-rAmylin binding could be displaced by CAL and amylin antagonists and agonists (Table 11). Fig.5 shows the effects of amylin displacement of 125I-rAmylin and125I-AC512. The low level of125I-rAmylin binding made rigorous quantification of this effect in COS-7 cells difficult.
Displacement of 125I-AC512 (•) and125I-rAmylin (○) by rAmylin in membranes from COS cells.[B o ], bound radioactivity. Log[Amylin], logarithms of molar concentrations of nonradioactive ligand. B, Data in A with ordinate values recalculated as a percentage of the initial BObinding of radioligand.
Receptor binding: HEK 293 cell membranes.
Saturable binding of the radioligands was observed in membranes from HEK 293 cells stably transfected with cDNA for hCTR2. The equilibrium dissociation constant of the 125I-AC512/receptor complex was 209 pm (95% CI, 133–331 pm; see Table12). The maximal number of binding sites (B max) was considerably larger than that found for COS-7 cell membranes (30,047 fmol/mg of protein; 95% CI, 24,130–35,964). Similar binding was observed with125I-sCAL (Kd = 195 pm; 95% CI, 56–692 pm) with a B max value not significantly different from that found with125I-AC512 (B max = 34,400 fmol/mg of protein; 95% CI, 24,063–44,737). Saturable125I-hCAL binding also was observed in these membranes (Kd = 219 pm; 95% CI, 151–316 pm). As was seen in COS-7 cell membranes, theB max value for125I-hCAL binding was significantly lower (6511 fmol/mg of protein; 95% CI, 3692–9329) than that seen with125I-AC512 and 125I-sCAL.
HEK 293 cells: saturation binding
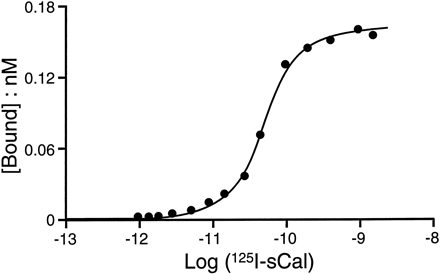
Membranes from HEK 293 cells also saturably bound125I-rAmylin (Kd = 49 pm; 95% CI, 28–89 pm). TheB max value for125I-rAmylin binding was again very much lower (2182 fmol/mg of protein; 95% CI, 1284–3080) than that found for125I-AC512 and 125I-sCAL and, notably, also that found for 125I-hCAL. The different B max values were studied further in HEK 293 cell membranes. Specifically, the saturation binding data for 125I-hCAL and125I-rAmylin were fit to a two-site (or two-state) model. A prerequisite to this procedure was a reliable estimate of the total number of binding sites. The saturation binding for 125I-sCAL was used since it clearly demonstrated a maximal asymptote for the saturation binding curve (Fig.6). It should be noted that although the dissociation kinetics of AC512 and hCAL were reversible,125I-sCAL gave essentially irreversible binding. Therefore, displacement of 125I-sCAL was an unsuitable method for receptor and/or ligand characterization. Specifically, the potency of displacing ligands and the magnitude of nonspecific binding of the pseudoirreversible ligand125I-sCAL was dependent on the binding protocol. Accordingly, if nonradioactive sCAL was added 30 min before the addition of 125I-sCAL, the observed IC50 was significantly lower than if the two ligands were added concomitantly to start the reaction. If the radioligand was added 30 min before the nonradioactive ligand, far less displacement was observed. Similar effects were observed with other nonradioactive ligands (i.e., hCAL). For these reasons, the use of125I-sCAL for receptor characterization was not pursued in these studies. However, the quantification of the maximal number of receptors with 125I-sCAL binding is still useful and, this was used to model the saturation binding curves for the agonist radioligands.
Saturation binding curve for 125I-sCAL to membranes from HEK 293 stably transfected with hCTR2 (clone 134).[Bound], Specific binding of 125I-sCAL.Log ( 125 I-cCal), logarithms of free molar concentrations of 125I-sCAL.
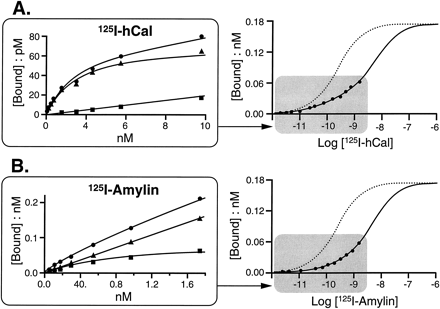
Fig. 7A (inset) shows the saturation curve for 125I-hCAL in membranes from stably transfected HEK 293 cells. The linear abscissal scale was transformed to a logarithmic scale and a fit to a model containing two affinity states for the maximal number of125I-sCAL binding sites (125I-sCAL saturation curve [dotted line]). For this particular experiment, the 125I-hCAL data indicated a 24.8% high affinity state. A similar procedure for the saturable binding for 125I-rAmylin (Fig. 7B) showed a significantly smaller number of high affinity sites (8.8% high affinity sites).
Saturation binding curves for125I-hCAL (A) and 125I-rAmylin (B) for hCTR2 (clone 134) expressed in HEK 293 cells. A, Inset, total (•), nonspecific (▪), and specific (▴) binding curves (inset is represented on larger axes as theshaded area). Log ( 125 I-hCal), logarithms of molar concentrations of 125I-hCAL. Dotted line, saturation binding curve for 125I-sCAL (see Fig. 6). The maximal asymptote of this latter curve was used to fit the binding data for 125I-hCAL to a two-site model. Under these conditions, the data for the specific binding of 125I-hCAL could be fit to a two-site model in which 24.8% of the sites had a high affinity for 125I-hCAL. B, Same as for A with125I-rAmylin as the radiolabel. The two-site model could be fit for an 8.8% population of high affinity sites for125I-rAmylin.
Bound radioligand could be displaced with agonists and antagonists for CTRs. The equilibrium dissociation constants of the nonradioactive displacing ligand/receptor complex (denoted as theKi ) for the displacement of the radioligands are given as negative log values (pKi ) in Table13. The pKi estimates for the displacement of all three radioligands with antagonists was uniform (Table 13); agonists had a higher estimated affinity for the displacement of125I-hCAL and 125I-rAmylin (as opposed to 125I-AC512).
HEK 293 cells: displacement of radiolabels
As was observed in COS-7 membranes, the curve for displacement of125I-hCAL was monophasic, whereas that for displacement of 125I-AC512 was biphasic and shifted to the right. In contrast, there was no significant difference in the potency of AC512 in displacement of these two radioligands. A similar effect, but more pronounced, was seen for amylin displacement of 125I-rAmylin and125I-AC512. Fig. 8shows the difference in potency demonstrated for amylin in displacement of these two radioligands.
Displacement of 125I-AC512 (•) and125I-rAmylin (○) by rAmylin in membranes from HEK 293 cells. A, B O, bound radioactivity. Log [hCal], logarithms of molar concentrations of nonradioactive ligand. B, Data recalculated as a percentage of the initial BO binding of radioligand.
Receptor binding: baculovirus expression.
Membranes fromTi ni cells infected with baculovirus containing cDNA for hCTR2 demonstrated saturable binding of125I-AC512 with aKd value of 575 pm (95% CI, 550–602 pm) and a B max value of 8340 fmol/mg of protein; 95% CI, 6,184–10,496). A comparable but somewhat larger number of sites was observed with 125I-sCAL (B max = 10,620 fmol/mg of protein; 95% CI, 9,208–12,031; Kd = 59 pm; 95% CI, 38–925 pm). In contrast, a significantly lower number of binding sites was observed for 125I-hCAL (Kd = 417 pm; 95% CI, 269–655 pm) with aB max value of 5460 fmol/mg of protein; 95% CI, 4,742–6,177). The data are summarized in Table14. No significant125I-rAmylin binding could be obtained in these membranes. Both agonist and antagonist ligands displaced125I-AC512 (see Table15 for pKi values).
Baculovirus expression: saturation binding
Baculovirus expression: displacement of radiolabels
Functional responses with hCTR2: microphysiometry.
In view of the complex displacement curves seen with some of these ligands and the submaximal saturation kinetics, it was useful to determine which of these ligands produced cellular responses (and thus could be classified as having efficacy with concomitant complex binding behavior). HEK 293 cells transfected with hCTR2 were tested in the cytosensor microphysiometer for the study of functional responses. A HEK 293 cell line expressing a high density of hCTR2 (B max = 28,000 fmol/mg of protein) yielded responses with complex wave forms that changed with hCAL concentration (Fig. 9A). Due to the rapidly declining phase of the response, dose-response curves could not be obtained in a cumulative manner (i.e., increases in concentration resulted in capricious secondary responses). Interestingly, a clone with a much lower receptor expression level (B max = 65 fmol/mg of protein; 95% CI, 40–96 fmol/mg of protein;Kd = 316 pm; 95% CI, 165–650 pm) provided an excellent functional response. In these cells, responses to hCAL were sustained (Fig. 9B) and yielded cumulative concentration-response curves (Fig.9C). AC512 produced concentration-dependent dextral displacement of the hCAL dose-response curve (Fig. 9D). Schild analysis with the antagonist AC512 provided a regression with a slope not significantly different from unity and a pKB value of 9.1 (95% CI, 9.48–8.7). This was not significantly different from the pKi or pKd value obtained in binding studies (see Tables 12 and 13).
Microphysiometry responses obtained from HEK 293 cells transfected with hCTR2. A. Response of a high receptor expression clone (clone C134–2-23; B max = 34, 400 fmol/mg of protein) to 10 nm hCAL. Percent, increase in hydrogen ion output as a percentage of the basal. B, Response of a low receptor expression clone (clone C134–4-7;B max = 67 fmol/mg of protein) to 10 nm hCAL. Percent, increase in hydrogen ion output as a percentage of the basal. C, Cumulative dose-response curve to hCAL. hCAL added at designated points a (10 pm), b (100 pm),c (1 nm), d (10 nm), and e (100 nm).Percent, increase in hydrogen ion output as a percentage of the basal. D, Dose-response curves to hCAL in the absence (•, six experiments) and presence of AC512 10 nm (○, three experiments), 100 nm (▪, three experiments), and 300 nm (▵, three experiments). Percent, responses as a percentage of the maximal response to 100 nmsCAL. Log [hCal], logarithms of molar concentrations of hCAL.
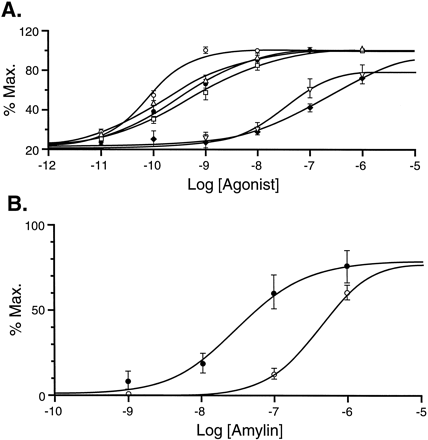
Receptor-transfected HEK 293 cells responded to a variety of agonists for CTRs and amylin receptors. Fig. 10A shows concentration-response curves to eel, porcine, and rat CAL; hCAL; rCGRP; and rAmylin. No responses to the antagonists AC66, AC413, hCGRP8–37, and AC512 were observed (data not shown). These data are in agreement with those obtained with binding, which showed that the observed affinity of the antagonists did not vary when displacing either the radioligand agonists (125I-hCAL or 125I-rAmylin) or antagonist (125I-AC512), whereas that of the agonists did. The dose-response curve to rAmylin was shifted to the right by AC512 (Fig. 10B). The resulting pA 2 value of 9.1 was not significantly different from the pKB estimated by Schild analysis for blockade of responses to hCAL.
Microphysiometry responses obtained from HEK 293 cells transfected with hCTR2 to agonists and antagonists for CTRs and amylin receptors. Max., increases in hydrogen ion output expressed as a percentage of the maximal output produced by 100 nm sCAL. Log, logarithms of molar concentrations of agonists. A, Responses to eel CAL (○, five experiments), hCAL (▵, five experiments), rat CAL (•, six experiments), porcine CAL (□, five experiments), rAmylin (▿, six experiments), and rCGRP (♦, six experiments). B, Blockade of amylin dose-response curve by AC512. Responses obtained in the absence (•, three experiments) and presence (○, three experiments) of AC512 (10 nm). Estimated pA 2 = 9.1.
Discussion
These data confirm the results reported previously by Beaumontet al. (6) that described the presence of a high affinity binding site for 125I-rAmylin in membranes prepared from rat nucleus accumbens. High affinity binding for the radiolabeled amylin receptor antagonist125I-AC512 also was confirmed, as reported previously by Watson et al. (9). The data for MCF-7 cell binding raise the question of the classification of the MCF-7 site as an operational human amylin receptor. This can be done by comparing the binding of ligands to the gene products expressed in the recombinant systems with the characteristics of amylin receptors in the natural systems, namely, the rat nucleus accumbens and MCF-7 cells. There are four unique characteristics of ligand binding to amylin receptors: (i) the high affinity binding for both 125I-rAmylin and 125I-AC512, (ii) the complex displacement kinetics between nonradioactive agonist amylin and antagonist radioligand 125I-AC512, (iii) the characteristically high affinity of rCGRP, and (iv) the similar affinities of the antagonists AC413, AC512, and AC66 in the rat nucleus accumbens and MCF-7 cells. In general, the binding with125I-AC512 and 125I-amylin showed these characteristics and, thus, support the notion that this site can be considered as an amylin receptor. However, the fact that hCGRP8–37 clearly distinguishes the rat and human receptor underscores the importance of species differences between the two receptors.
The original intent of this study was to define the human high affinity receptor for amylin in MCF-7 cells. Expression cloning from a library prepared from the human breast carcinoma MCF-7 cell line with a radiolabeled form of the amylin and CAL peptide antagonist AC512 yielded two hCTR isoforms, hCTR1 and hCTR2 (10, 15-17). The presence of these receptor isoforms in this cell line has been demonstrated by Albrandt et al. (18) through reverse transcription-polymerase chain reaction amplification from reported sequences. The hCTR1, first described in BIN 67 cells (15), saturably bound 125I-AC512 and125I-sCAL. It is interesting to note that the affinity of 125I-sCAL is considerably higher in the current membrane study than that reported for whole-cell binding [estimated pKi = 10 (16) to 8.0 (19)]. The higher affinity reported in this study may reflect receptor isomerization due to formation of G protein complexes, a common difference between membrane and whole-cell binding studies; however, it is difficult to interpret estimated affinities with this ligand because the high affinity for the receptor also may reflect the pseudoirreversible kinetics of onset. The unsuitability of125I-sCAL in receptor characterization (other than for quantification of receptor number) provided the impetus to characterize the affinities to the agonist and antagonist ligands with125I-AC512. The lack of high affinity displacement by rAmylin and the lack of saturable binding for125I-rAmylin precluded further exploration of a possible relationship between hCTR1 and amylin in this study; in contrast, the high affinity binding of125I-rAmylin to expressed hCTR2 provided a basis for further study. As a prerequisite to a discussion of the data with hCTR2, it is useful to delineate the data in terms of those obtained with agonists and those obtained with antagonists. Ligands with no efficacy are very useful in the process of expression cloning procedure and subsequent identification of products. Unlike those of agonists, the observed binding affinities of antagonists do not vary because of receptor/G protein complexation. The studies with the microphysiometer were helpful in classifying the ligands used in this study; the microphysiometry data with these ligands indicated AC512, AC66, AC413, and hCGRP8–37 produced no visible response and, thus, ostensibly qualified as antagonists. By implication,125I-AC512 also was considered to be a nonefficacious antagonist. The affinity of this radioligand from saturation binding varied <2.5-fold among the various expression systems, indicating that the environment of the gene product did not appreciably affect the binding of this ligand. Similarly, the affinity of the antagonist AC413 in displacing the radioligand antagonist125I-AC512 was constant over the three expression systems (COS-7, HEK 293, and Ti ni cells). However, some variation for AC66 (Ti ni cells) and hCGRP8–37 was observed. In general, the data suggest that the hCTR2 antagonist binding was fairly consistent in the various expression systems used in this study.
A recurrent finding in this work was that agonists were more potent at displacing radiolabeled agonists (125I-hCAL,125I-rAmylin) than they were at displacing the radiolabeled antagonist 125I-AC512. This is consistent with the idea that the agonist radioligands select the high affinity species in a G protein-deprived environment, whereas the antagonist labels a random sampling of bare receptors. The concept of G protein deprivation does not necessarily refer to a stoichiometric deficiency in the ratio of expressed receptors to G proteins but rather to an inability of the expressed receptors to adequately access the existing G proteins due to constraints in the membrane architecture (20). On displacement with nonradioactive agonist, insufficient G protein exists for complete formation of the high affinity ternary complex; thus, a lower affinity (agonist binding to the receptor not complexed with G protein) is observed. This effect of high affinity selection is made manifest in the significantly different pKi values for agonists when displacing agonist and antagonist radioligands. The idea that there is a G protein insufficiency in some of these systems is supported by the significantly lower B max values for radioactive agonists versus the antagonist125I-AC512. The data for125I-hCAL indicate that COS-7 cells and baculovirus-expressed Ti ni cells have an equal capability to form the high affinity ternary complex for hCAL (68% and 65% of the receptors, respectively), whereas HEK 293 cells are limited (only 22%). In all cases, however, the maximal number of binding sites measured by agonist saturation binding was much less than the number of receptors as quantified by binding of the antagonist125I-AC512. The observance of differences in apparent receptor densities when measured with agonist and antagonist radioligands is known (21). However, differences in the potencies of agonist ligands when displacing agonist and antagonist radioligands usually produce complex biphasic displacement curves for antagonist displacement. The production of parallel displacement curves with Hill coefficients near unity is more uncommon but also not unknown (22).
In general, the experimental results can be discussed in terms of two possible hypotheses. The first is that amylin is simply a low efficacy agonist for the CTR. Under these circumstances, the hCTR2 clone is not associated with the amylin binding found in MCF-7 cells (i.e., the amylin receptor gene product was not recovered from these studies). The second is that the CTR functions as the amylin receptor when coupled to certain G proteins in some membranes. These ideas are considered separately.
The first hypothesis to consider is the proposition that amylin is simply a lower efficacy agonist for hCTR2 in these systems and that the amylin effect is not relevant to the amylin binding seen in native MCF-7 cells. In terms of this idea, the two gene products isolated from the MCF-7 cell library are not related to the high affinity amylin binding found in the MCF-7 cells. The small quantity of high affinity binding of 125I-rAmylin found in COS-7 and HEK 293 cells and the high affinity selection effects would then represent a separate activity of rAmylin for the hCTR2. In terms of this hypothesis, a human amylin receptor awaits discovery in the MCF-7 cell library. This hypothesis is based on the interaction between receptors and G proteins. The ternary complex models for seven-transmembrane receptors incorporate an intrinsic affinity constant between the activated receptor and G proteins (23-26). Under these circumstances, there is a stoichiometric relationship between the receptor and G protein that depends on this affinity constant and the relative amounts of receptor and G protein. In G protein-deprived systems, the amount of activated receptor dictates the amount of complex of receptor and G protein. Thus, a low efficacy partial agonist may produce a lower amount of activated receptor than a high efficacy agonist with a subsequently lower quantity of high affinity agonist complex. These effects have been observed directly with cholinergic agonists (27, 28) (see Ref. 29 for a discussion). The fact that hCAL produces 24.8% high affinity complex in HEK 293 cells while amylin produces only 8% is consistent with the idea that amylin produces less of the activated state than hCAL (i.e., it simply is a lower efficacy agonist for the CTR).
A second hypothesis describes a system in which hCTR2 couples to one G protein for CAL function and another for amylin function. This latter G protein may not be present in all cellular systems, thus conferring cellular selectivity for amylin effect. There is a considerable body of evidence to show that CTRs in general couple to a variety of G proteins [i.e., Gs, Gi, and Gq; see Horne et al. (30) for a review]. In addition, the promiscuous coupling of this receptor has been shown to alter with cell cycle as well (31). In the current series of experiments, there are two lines of evidence consistent with a hypotheses of separate G protein coupling for hCAL and rAmylin. The first is the disparate formation of high affinity ternary complexes formed by hCAL and rAmylin as measured by radioligand saturation binding in all of these systems (i.e., 24.8% for125I-hCAL versus 8% for125I-rAmylin). In terms of this hypothesis, the different B max values for the two agonists may reflect different stoichiometries of the different G proteins. However, this is not definitive because this also is consistent with amylin simply being a lower efficacy agonist for hCTR2 (see above).
The main support for the second hypothesis is related to the extremely selective affinity of rAmylin and rCGRP for displacement of125I-rAmylin and the considerably lower potency in displacing 125I-hCal (i.e., they are of low activity at CTRs). This suggests that the CTR changes character when bound to 125I-amylin. In view of the high degree of high affinity selection for 125I-rAmylin displacement over displacement of 125I-AC512 observed for amylin in MCF-7 cells (the source of the transfected receptor hCTR2) and the fact that the same is not true for hCAL suggest that the MCF-7 cell possesses a G protein or other factor to confer high rAmylin binding in that system and that this factor is lost on transfection into COS-7, HEK 293, and Ti ni cells. The factor need not be a specific G protein; it could be an auxiliary player in the receptor coupling process. For example, protein factors that tightly couple adenosine receptors (32) and α2A/D-adrenergic receptors (33) have been described recently. Removal of these from host membranes results in loss of high affinity binding of agonists to receptors.
In conclusion, these data describe receptor binding characteristics of two hCTR isoforms in various expression systems for CAL and amylin ligands. The provocative association of the hCTR2 with high affinity amylin binding requires further study and may have implications for the understanding of selective cellular signaling of hormones and the use of gene products to attain signaling diversity. In view of the similar affinities of all of these agonist and antagonist radioligands for amylin binding in MCF-7 cells and that found in these expression studies with hCTR2, it could be inferred that these effects are mediated either by one receptor coupling to various membrane components or by separate but similar receptors.
Footnotes
- Received February 26, 1997.
- Accepted July 28, 1997.
-
Send reprint requests to: Dr. Terry P. Kenakin, Department of Receptor Biochemistry, Glaxo Wellcome Inc., 5 Moore Drive, Research Triangle Park, NC 27709.
Abbreviations
- rAmylin
- rat amylin
- CTR
- calcitonin receptor
- hCTR
- human calcitonin receptor
- HEK
- human embryonic kidney
- HEPES
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- sCAL
- salmon calcitonin
- hCAL
- human calcitonin
- Ti ni
- Trichoplusia ni
- rCGRP
- rat calcitonin gene-related product
- hCGRP
- human calcitonin gene-related product
- CI
- confidence interval
- sCAL
- salmon calcitonin
- hCAL
- human calcitonin
- The American Society for Pharmacology and Experimental Therapeutics