Abstract
Aryl hydrocarbon receptor (AhR) is a ligand-mediated transcription factor known for regulating response to xenobiotics, including prototypical 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the activation of CYP1A1 expression. Upon ligand-binding, AhR translocates to the nucleus, interacts with the AhR nuclear translocator, and binds to xenobiotic response elements (XREs; GCGTG) present in the promoter region of AhR-regulated genes. Recently, we identified a novel tryptophan catabolite, cinnabarinic acid (CA), as an endogenous AhR agonist capable of activating expression of AhR target gene stanniocalcin 2 (stc2). The CA-driven stc2 induction bestowed cytoprotection against hepatotoxicity in an AhR-dependent manner. Interestingly, only CA but not TCDD was able to induce stc2 expression in liver, and CA was unable to upregulate the TCDD responsive cyp1a1 gene. In this report, we identified CA-specific histone H4 lysine 5 acetylation and H3 lysine 79 methylation at the AhR-bound stc2 promoter. Moreover, histone H4 lysine 5 acetylation writer, activating transcription factor 2 (Atf2), and H3 lysine 79 methylation writer, disruptor of telomeric silencing 1-like histone lysine methyltransferase (Dot1l), were interacting with the AhR complex at the stc2 promoter exclusively in response to CA treatment concurrent with the histone epigenetic marks. Suppressing Atf2 and Dot1l expression using RNA interference confirmed their role in stc2 expression. CRISPR/Cas9–assisted replacement of cyp1a1 promoter–encompassing XREs with stc2 promoter XREs resulted in CA-dependent induction of cyp1a1, underlining a fundamental role of quaternary structure of XRE sequence in agonist-specific gene regulation. In conclusion, CA-driven recruitment of specific chromatin regulators to the AhR complex and resulting histone epigenetic modifications may serve as a molecular basis for agonist-specific stc2 regulation by AhR.
SIGNIFICANCE STATEMENT Results reported here provide a mechanistic explanation for the agonist-specific differential gene regulation by identifying interaction of aryl hydrogen receptor with specific chromatin regulators concomitant with unique histone epigenetic marks. This study also demonstrated that the agonist-specific target-gene expression can be transferred with the gene-specific promoter xenobiotic response element-sequence in the context of chromatin architecture.
Introduction
The basic helix-loop-helix period circadian protein/aryl hydrocarbon receptor nuclear translocator (Arnt)/single-minded protein domain family (βHLH/PAS) of transcription factors and regulators have distinct physiologic, pathologic, and developmental functions despite conserved domain architecture (McIntosh et al., 2010). Within the family, the aryl hydrocarbon receptor (AhR) is a key transcription factor activated by a number of xenobiotics, including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Nebert and Gelboin, 1968a,b; Legraverend et al., 1982; Hankinson, 1993, 1995; Nebert et al., 1993). In an unliganded state, AhR resides in the cytoplasm in complex with molecular chaperonins—heat shock protein 90, p23, and AhR-interacting protein (Ma et al., 2009; Flaveny et al., 2010). Upon TCDD binding, AhR dissociates from chaperonins, translocates to the nucleus, and heterodimerizes with Arnt (Fukunaga et al., 1995). The liganded AhR-Arnt complex then binds to xenobiotic response elements (XREs, GCGTG motif) present in the promoter region of AhR target genes, including archetypical cyp1a1 (Jones et al., 1986; Elferink et al., 1990; Elferink and Whitlock, 1990).
Since the discovery of AhR, a wide range of xenobiotics, including polycyclic aromatic hydrocarbons, halogenated aromatic hydrocarbons, and polychlorinated biphenyls, have been identified as exogenous AhR ligands (Nguyen and Bradfield, 2008). Additionally, recent advancement in the field included a plethora of structurally diverse bacterial products and dietary and endogenous compounds as AhR agonists (Denison and Nagy, 2003; Denison et al., 2011; Hubbard et al., 2015). Among them, cinnabarinic acid (CA), a tryptophan metabolite and byproduct of the kynurenine pathway, has been shown to activate AhR (Lowe et al., 2014). Upon CA treatment, the AhR-Arnt complex is directly recruited to the eight XREs clustered in a 218-bp region of the stanniocalcin 2 (stc2) promoter (Harper et al., 2013). Interestingly, in hepatocytes, only CA but not TCDD induced stc2 expression through an XRE-driven mechanism, whereas CA, in contrast to TCDD, did not upregulate cyp1a1 expression (Harper et al., 2013; Joshi et al., 2015). The CA-driven AhR-dependent stc2 upregulation was responsible for the protection against endoplasmic reticulum/oxidative stress–induced apoptosis both in vitro and in vivo (Joshi et al., 2015). To investigate the molecular basis for the agonist-specific, mutually exclusive transcription response, we previously employed mass spectrometry on immunoaffinity purified AhR complexes captured after CA or TCDD treatments (Joshi et al., 2017). Mass spectrometry identified CA-specific interaction of AhR with metastasis-associated protein 2 (Mta2), a known chromatin regulator, concomitant with histone H4 lysine 5 acetylation (H4 K5ac). Moreover, H4 K5ac was absolutely dependent on CA-induced AhR-Mta2 recruitment to the stc2 XREs and played critical role in the regulation of stc2 gene expression (Joshi et al., 2017). The current study extends our previous observation and uses crosslinking chromatin immunoprecipitation–coupled mass spectrometry (xChIP-MS) to identify additional histone epigenetic marks, namely TCDD-specific histone H3 lysine 14 acetylation (H3 K14ac), histone H3 lysine 23 acetylation (H3 K23ac), and histone H3 lysine 27 dimethylation (H3 K27dime) as well as CA-exclusive histone H3 lysine 79 methylation (H3 K79me) at the AhR-bound chromatin complex.
In the present study, we demonstrated transient binding of H4 lysine 5 acetylation and stable association of H3 K79me at the AhR-bound stc2 promoter in response to CA treatment. Moreover, H4 K5ac and H3 K79me marks were concurrent with the interaction of histone modification writers activating transcription factor 2 (Atf2) and disruptor of telomeric silencing 1-like histone lysine methyltransferase (Dot1l) to the AhR complex at the stc2 promoter resulting in target-gene induction. Finally, this study has enhanced our understanding of AhR biology by exhibiting that the dynamic quaternary structure of the stc2 promoter containing XREs encodes comprehensive epigenetic and chromatin structural information necessary for the CA-specific AhR binding and AhR-mediated transcription of stc2.
Materials and Methods
Animals, Cell Culture, and Treatments
Eight- to ten-week-old C57BL/6 (wild-type, WT) female mice (Jackson Laboratories, Bar Harbor, ME) were used in compliance with the guidelines of the Institutional Animal Care and Use Committee at the University of Oklahoma Health Sciences Center and the University of Texas Medical Branch. Mice were treated by oral gavage with vehicle (corn oil), 20 µg/kg TCDD (AccuStandard, New Haven, CT), or intraperitoneal injection with 12 mg/kg CA (synthesized by Synthetic Organic Chemistry Core at University of Texas Medical Branch) for 0, 5, 10, 15, and 30 minutes and 1, 2, 4, 6, 8, 12, 24, and 48 hours before sacrifice. For cell culture experiments, alpha mouse liver 12 (AML12) cell line, a differentiated nontransformed mouse hepatic cells (CRL-2254; American Type Culture Collection, Manassas, VA) were plated at a density of 500,000 cells/cm2 in Dulbecco’s modified Eagle’s medium:F12 medium containing 10 µg/ml insulin, 5.5 µg/ml transferrin, 5 ng/ml selenium, 40 ng/ml dexamethasone, 100 U/ml penicillin, 100 µg/ml streptomycin, and 5% FBS. AML12 cells were transiently transfected with ON-TARGETplus Atf2 and Dot1l small interfering RNAs (siRNAs) (Thermo Fisher Scientific, Waltham, MA) for 24 hours using Metafectene PRO transfection agent (Biontex Laboratories, GmbH, München, Germany). Cells were further treated with 6 nM TCDD, 30 µM CA, or vehicle (DMSO) for 2 hours. In both cell culture and animal studies, treatment with vehicle, TCDD, and CA were performed blindly to the experimenter by another individual.
xChIP-MS
Upon TCDD or CA treatments, liver tissues from WT mice were extracted, finely minced, and subjected to two-step crosslinking chromatin immunoprecipitation (Tian et al., 2012). Briefly, minced livers were crosslinked using 2 mM disuccinimidyl glutarate (DSG, 7.7 Å spacer arm) (Thermo Fisher Scientific) in phosphate-buffered saline for 45 minutes at room temperature. Further crosslinking with 1% formaldehyde (Thermo Fisher Scientific) in phosphate-buffered saline was carried out at room temperature for 10 minutes. Crosslinked samples were homogenized using Dounce homogenizer and centrifuged at 3200 × g for 5 minutes at 4°C. The supernatant was discarded, and the pellet was resuspended in the cell lysis buffer (150 mM NaCl, 25 mM Tris [pH 7.5], 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, and protease inhibitor cocktail), Dounce homogenized, incubated on ice for 20 minutes, centrifuged at 3200 × g for 5 minutes at 4°C, and processed using ChIP-IT Express Shearing Kit (Active Motif, Carlsbad, CA). Genomic DNA shearing was performed with Adaptive Focused Acoustics sonicator (Covaris, Woburn, MA) to yield ≈ 400-bp DNA fragments bound to the protein complex. Immunoprecipitation was carried out by antibody-targeting AhR (Abcam, Cambridge, MA). Protein-DNA complexes were eluted using elution buffer provided in the kit. Proteins were extracted using SDS loading buffer containing 100 mM dithiothreitol, which was followed by incubation at 100°C for 10 minutes. Proteins were separated using SDS-PAGE gel electrophoresis, stained with Coomassie Blue, destained, and had their bands cut out for subsequent mass spectrometry analysis. Gel bands were washed three times with 50% methanol and deionized water, dried by a piece of tissue paper, and ground into fine powder with a tip-sealed 200-µl pipette tip. Fifty millimolar ammonium bicarbonate (100 µl) was added to cover the gel powder. Samples were digested overnight at 37°C by addition of 2 µg trypsin. Digested peptides were extracted by acetonitrile, dried by speedvac, and then redissolved in 50 µl of 1% formic acid for liquid chromatography with tandem mass spectrometry analysis. Peptide mixtures were separated by reversed-phase liquid chromatography using an Easy-nLC equipped with an autosampler (Thermo Fisher Scientific). A PicoFrit 25-cm length × 75-µm internal diameter, ProteoPep analytical column packed with a mixed (1:1) packing material (Waters XSelect HSS T3, 5 µ, and Waters YMC ODS-AQ, S-5, 100Å) was used to separate peptides by reversed-phase liquid chromatography (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile), which ran with a 176-minute gradient from 2%–45% of solvent B with a flow rate at 300 µl/min. The QExactive mass analyzer (Thermo Fisher Scientific GmbH, Bremen, Germany) was set to acquire data at a resolution of 35,000 in full scan mode and 17,500 in tandem mass spectrometry (MS/MS) mode. The top 15 most intense ions in each mass spectrometry survey scan were automatically selected for MS/MS. Peptides were identified by PEAK 8.5 (Bioinformatics Solutions, Waterloo, Canada) to perform de novo sequencing assisted search against the mouse database (Searched Entry: 52485). Acetylation, monomethylation, dimethylation, trimethylation, and citrullination of lysine were set as variable modifications. False discovery rates were estimated by the ratio of decoy number of hits over target number of hits among peptide spectrum matches. The maximum allowed −10logP is ≥15.
RNA Isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction
Total RNA was isolated from vehicle-, TCDD-, and CA-treated mice livers and AML12 cells using Trizol (Thermo Fisher Scientific). cDNA was prepared from 1 µg total RNA using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed using StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using cyp1a1 Forward 5′-GCCTAACTCTTCCCTGGATGC-3′, cyp1a1 Reverse 5′-TCAATGAGGCTGTCTGTGATGTC-3′, stc2 Forward 5′-GTCGGTGTGATTGTGGAGATGAT, stc2 Reverse 5′-TCCACATAGGGCTCATGCAG, 18S ribosomal RNA (rRNA) Forward 5′-CTCAACACGGGAAACCTCAC-3′, and 18S rRNA reverse 5′-CGCTCCACCAACTAAGAACG-3′ primers and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific).
Chromatin Immunoprecipitation
After treatments with vehicle, TCDD, and CA, liver tissues from WT mice were extracted, finely minced, and fixed with 1% formaldehyde in phosphate-buffered saline at room temperature for 10 minutes. Livers were homogenized using Dounce homogenizer, centrifuged at 3200 × g for 5 minutes at 4°C, and resuspended in 2 ml cell lysis buffer (150 mM NaCl, 25 mM Tris [pH 7.5], 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, and protease inhibitor cocktail). Samples were incubated on ice for 15 minutes, centrifuged at 3200 × g for 5 minutes at 4°C, and processed through ChIP-IT Express Enzymatic Kit (Active Motif) according to the manufacturer’s instructions. AML12 cells were crosslinked with 1% formaldehyde and processed as per the manufacturer’s protocol (Active Motif). Antibodies against AhR and histone H3 (Abcam); histone H4, H4 K5ac, H3 K79me, and IgG (Cell Signaling Technology, Danvers, MA); and Atf2 and Dot1l (Santacruz Biotechnology, Dallas) were used to immunoprecipitate protein-bound DNA complexes. Immunoprecipitated and input DNA was polymerase chain reaction (PCR)-amplified using primers specific to the cyp1a1 and stc2 promoters flanking the XREs. The cyp1a1 and stc2 PCR primer pairs are cyp1a1 forward 5′-CTATCTCTTAAACCCCACCCCAA-3′, cyp1a1 reverse 5′-CTAAGTATGGTGGAGGAAAGGGTG-3′, stc2 forward 5′-CTCAGTCCATTCGGCCATTGCCC-3′, and stc2 reverse 5′-AGGAAGCGGAGCGCCTCCGC-3′. For chromatin immunoprecipitation (ChIP) assays performed on the CRISPR/Cas9 edited AML12 cells, cyp1a1 forward 5′-CAGGGGAGGGCAGGTGAAGG-3′ and cyp1a1 reverse 5′-TGGTGACTTTGCTTCCCTGG-3′ primers were used. PCR products were fractionated on a 5% polyacrylamide gel, stained with SYBR Green (Thermo Fischer Scientific), and imaged on a ChemiDoc MP imaging system (Bio-Rad).
Crosslinking Chromatin Immunoprecipitation–Western Blotting and Western Blotting
Two-step crosslinking chromatin immunoprecipitation (xChIP) using anti-AhR antibody was performed as described in the aforementioned xChIP-MS section. Upon crosslinking and chromatin immunoprecipitation using anti-AhR antibody, proteins were extracted with SDS loading buffer containing 100 mM dithiothreitol, which was followed by incubation at 100°C for 10 minutes. Protein samples were fractionated by SDS-PAGE using the Mini-Protean electrophoresis system (Bio-Rad) and transferred to 0.45-µm low-fluorescence polyvinylidene difluoride membranes (Bio-Rad) using Trans-Blot Turbo system (Bio-Rad). Membranes were probed with antibodies against histone H4, H4 K5ac, H3 K79me, H3 K14ac, H3 K23ac, H3 K27dime, histone acetyltransferase p300 (p300), and CREB-binding protein (Cbp) (Cell Signaling Technology); Atf2, Dot1l, histone acetyltransferase Tip60 (Tip60), Mta2 (Santacruz Biotechnology); and histone H3 (Abcam). For AML12 cells, extracts were prepared using cell lysis buffer (Cell Signaling Technology) and upon Western blotting, were probed with anti-Atf2, anti-Dot1l (Santacruz Biotechnology), and anti-actin (Millipore Sigma, St. Louis, MO) antibodies. Proteins were detected using IRDye 800CW and IRDye 680RD secondary antibodies (Li-COR, Lincoln, NE), which was followed by imaging using an Odyssey CLx imaging system (Li-COR).
Replacement of cyp1a1 Promoter XREs with “stc2 XRE Cassette” in AML12 Cells
Using CRISPR/Cas9 editing, a modified AML12 cell line was constructed by replacing a 926-bp cyp1a1 promoter region (between −574 and −1500 from the transcription start site) containing 10 XREs with a 259-bp stc2 promoter region (−210 to −469) encompassing eight XREs—“stc2 XRE cassette” (Supplemental Fig. 1A). Using Universal CRISPR activity assay (Biocytogen, Wakefield, MA), TCTGGGCTCGGGAGCTCACA GGG as 5′ single guide RNA and GGCACCCATTGGCTTGTAGT AGG as 3′ single guide RNA were chosen. Target vector construction, electroporation, and screening of positive clones were performed (Biocytogen). Junction PCR using homologous recombination (HR) allele primer pair CL-JGY-002-A-L-GT-F (TGTAAGGGTCGGGTGTCCTGAGAAT) and Puromycin-GT-F (GCAACAGATGGAAGGCCTCCTGGCG), HR allele primer pair CL-JGY-002-A-R-GT-F (GCGGTCCTTCGGGCACCTCGAC) and CL-JGY-002-A-R-GT-R (TGGTGTTTCCAGTTCCCTGAAGCTC), non-HR allele primers CL-JGY-002-A-R-GT-F1 (GTTGTAAACTGTCCCCTGCATATC) and CL-JGY-002-A-R-GT-R (TGGTGTTTCCAGTTCCCTGAAGCTC), and DNA sequencing confirmed successful donor vector integration. A homozygous clone D02 (referred to as CRISPR/Cas9-edited AML12 cells) was used in the present studies (Supplemental Fig. 1).
Statistical Analysis
xChIP-MS and xChIP–Western blotting assays to identify TCDD- and CA-specific histone modifications and modifiers, respectively, were exploratory in nature. The xChIP–Western blotting (to confirm newly identified histone modifications), ChIP, and quantitative RT-PCR experiments were conducted with a preset plan. The sample sizes per group, blinding, and data analysis methodology were predetermined (Michel et al., 2020). Data were analyzed by applying ANOVA models using Sigma Stat software (Systat Software, San Jose, CA). Differences between the groups were considered significant only if the P value was <0.05.
Results
CA and TCDD Induced Differential Gene Expression of stc2 and cyp1a1
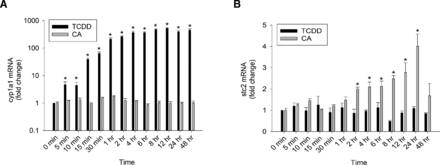
We have previously assessed mutually exclusive expression of AhR target genes, cyp1a1 and stc2, in response to 2 hours of TCDD and CA treatments (Joshi et al., 2015, 2017). Here, we measured cyp1a1 and stc2 mRNA message in the livers of TCDD-treated (20 µg/kg) and CA-treated (12 mg/kg) WT mice at different time points (Fig. 1). TCDD-driven induction of cyp1a1 was observed at 5 minutes and plateaued at 1 hour (Fig. 1A). Maximal stc2 induction by CA was achieved at 24 hours with significant reduction in the message at 48 hours (Fig. 1B). Vehicle treatment did not elicit cyp1a1 or stc2 induction (Supplemental Fig. 2A). The data confirmed that the agonist-specific dichotomous expression of cyp1a1 and stc2 persists temporally.
Agonist-specific dichotomous expression of stc2 and cyp1a1. WT mice were treated with 20 µg/kg of TCDD and 12 mg/kg of CA for the indicated time. RT-PCR was performed to quantitate (A) cyp1a1 and (B) stc2 mRNA levels in liver normalized against 18S ribosomal RNA. For statistical analysis, a mixed-effects multivariate ANOVA (MANOVA) model was used. After an overall significant F test from the MANOVA model, the post hoc multiple-comparison tests were performed for the prespecified comparisons adjusted by Tukey procedure. *P < 0.05, and n = 3 independent mice.
Identification of CA- and TCDD-Specific Histone Post-Translational Modifications
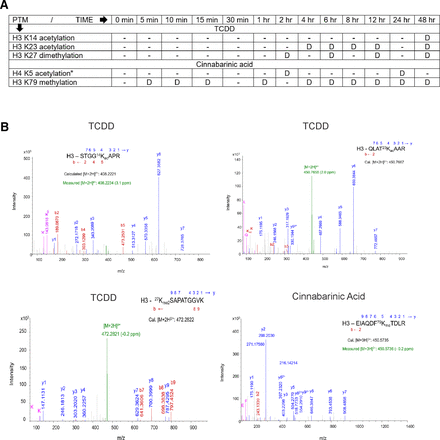
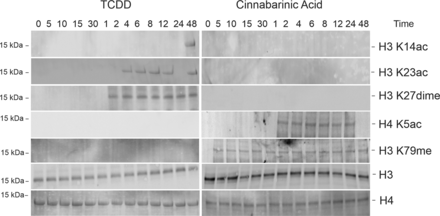
To evaluate involvement of distinct epigenetic modifications in agonist-specific differential gene regulation, CA- and TCDD-treated livers were crosslinked with protein-protein and protein-DNA crosslinkers and subjected to xChIP-MS (Tian et al., 2012; Sowers et al., 2015). xChIP-MS identified TCDD-specific H3 K14ac, H3 K23ac, and H3 K27dime and CA-specific stable H3 K79me marks at the AhR-bound chromatin complex (Fig. 2, A and B). Moreover, we have previously confirmed association of H4 K5ac at the AhR-bound stc2 promoter after CA treatment but not TCDD or vehicle treatment (Fig. 2A) (Joshi et al., 2017). To verify interaction of specific epigenetic marks within AhR-bound chromatin, xChIP–Western blotting was performed. TCDD-specific association of the AhR complex with H3 K14ac at 48 hours; H3 K23ac at 4, 6, 8, 12, and 48 hours; and H3 K27dime between 2 and 48 hours was validated (Fig. 3). H3 K14ac, H3 K23ac, and H3 K27dime marks were not observed at AhR-bound chromatin upon CA or vehicle treatment (Fig. 3; Supplemental Fig. 2B). CA-specific, AhR-bound H4 K5ac expression was transient between 2 and 24 hours, which was contemporaneous with CA-induced stc2 expression (Fig. 3). xChIP-immunoblotting also confirmed stable interaction of H3 K79me with CA-induced AhR complex (Fig. 3).
Identification of CA- and TCDD-specific, AhR-associated epigenetic modifications. WT mice were gavaged with 20 µg/kg of TCDD or intraperitoneally injected with 12 mg/kg of CA for the denoted time period. Two-step crosslinking chromatin immunoprecipitation was performed on livers using anti-AhR antibody followed by liquid chromatography coupled tandem mass spectrometry. (A) summarizes mutually exclusive histone modifications detected (denoted by D) upon TCDD and CA treatments at various time-points. * denotes H4 K5 acetylation previously detected by mass spectrometry and Western blotting after 2- and 24-hour CA treatment. (B) Representative high-resolution MS/MS spectra of STGG14KacAPR (encompassing residues 10–17), QLAT23KacAAR (residues 19–26), 27Kme2SAPATGGVK (residues 27–36) and EIAQDF79KmeTDLR (encompassing residues 73–83) in histone H3. For CA-specific H4 K5 acetylation, high-resolution spectra of G5KacGGKGLGKGGAKR (encompassing residues 4–17 in histone H4) with detailed information regarding theoretical and observed mass-to-charge ratio (m/z) values for fragment ions were published previously (Joshi et al., 2017).
Histone H4 K5 acetylation and H3 K79 methylation at the AhR-chromatin complex exclusively upon CA treatment. Crosslinked chromatin immunoprecipitated (with anti-AhR antibodies) protein extracts were subjected to Western blotting and probed with anti-histone modification antibodies. One representative blot is shown (n = 3 independent mice). Histone H3 and H4 were used as loading controls. Times 0, 5, 10, 15, and 30 indicate minutes and 1, 2, 4, 6, 8, 12, 24, and 48 are hours of 20 µg/kg of TCDD and 12 mg/kg of CA treatment.
Chromatin Regulators Associated with AhR-Bound Chromatin Complex
Given that the histone post-translational modifications are vital for regulating chromatin architecture, and dynamic homeostasis of these modifications are driven by the recruitment of chromatin regulators such as “writers” of histone modifications (Gillette and Hill, 2015), we sought to identify AhR-bound regulators of CA-specific histone modifications. We focused on specific histone modification “writers” that are known or are likely to trigger histone H4 lysine 5 acetylation (H4 K5ac) and histone H3 lysine 79 methylation (H3 K79me) (Marmorstein and Zhou, 2014; Sandoval et al., 2016; Hyun et al., 2017). xChIP–Western blotting revealed CA-specific interaction of known H4 K5 acetylation writer Atf2 with AhR-chromatin complex between 2 and 24 hours, which was concurrent with H4 K5 acetylation and stc2 induction (Fig. 4; Supplemental Fig. 2C). We were able to recapitulate previously detected association of histone reader Mta2 with AhR uniquely in response to CA treatment (Joshi et al., 2017). Furthermore, CA treatment resulted in an interaction of AhR-bound chromatin complex with a known H3 K79 methylation writer, Dot1l (Fig. 4). Other lysine acetyltransferases examined, including p300, Cbp, and Tip60, were associated with AhR-chromatin complex in response to both TCDD and CA treatments but not upon administration of the vehicle (Fig. 4 and Supplemental Fig. 2C).
Identification of AhR-associated chromatin modification writers of H4 K5 acetylation and H3 K79 methylation. Livers of TCDD- and CA-treated WT mice were chromatin immunoprecipitated with anti-AhR antibody as described in Materials and Methods. Immunoblotting was carried out to detect enrichment of known histone modification writers of H4 K5 acetylation and H3 K79 methylation. Western blots shown are representative results from three independent experiments. 0, 5, 10, 15, and 30 indicate time in minutes and 1, 2, 4, 6, 8, 12, 24, and 48 are hours of 20 µg/kg of TCDD and 12 mg/kg of CA treatment.
CA-Specific Direct Recruitment of Chromatin Modification Writers to stc2 Promoter In Vivo
The 218-bp region of the stc2 promoter between −244 and −462 bp upstream of the transcription start site contains eight distinct XREs (Harper et al., 2013). Prior studies have demonstrated recruitment of the AhR-Arnt-Mta2 complex to the stc2 promoter sequence in response to CA treatment (Joshi et al., 2015, 2017). Here, we examined whether histone H4 K5 acetylation and H3 K79 methylation writers, Atf2 and Dot1l, were recruited to the stc2 promoter encompassing the XRE cluster. Chromatin immunoprecipitation assay was performed on whole-liver tissue targeting XREs within the stc2 and cyp1a1 promoter regions. CA-specific recruitment of Atf2 to the stc2 promoter was observed between 2 and 24 hours, which was concurrent with the AhR binding (Fig. 5), H4 K5 acetylation (Fig. 3), and elevated stc2 expression (Fig. 1). Dot1l was stably bound to the stc2 promoter upon CA treatment (Fig. 5). Neither Atf2 nor Dot1l was recruited to the cyp1a1 promoter upon CA treatment or to the stc2 and cyp1a1 XREs upon TCDD or vehicle administration (Fig. 5; Supplemental Fig. 2D). Finally, AhR was bound directly to the cyp1a1 promoter upon TCDD treatment, which induced cyp1a1 expression (Fig. 5).
CA-dependent Atf2 and Dot1l binding to the stc2 promoter in vivo. ChIP assays were performed on livers from WT mice treated with TCDD (20 µg/kg) and CA (12 mg/kg) for 0, 5, 10, 15, and 30 minutes and 1, 2, 4, 6, 8, 12, 24, and 48 hours. Antibodies against the histone modification writers Atf2 and Dot1l and against AhR were used to immunoprecipitate the target proteins. Anti-IgG and anti-H3 antibodies were used as negative and positive controls, respectively. PCR using primers targeting XRE clusters in the stc2 and cyp1a1 promoters were used to amplify the precipitated DNA. PCR products were separated on two 5% polyacrylamide gels and visualized with SYBR Green dye. Samples were run, stained with SYBR Green, and imaged on Chemidoc MP (Bio-Rad) simultaneously with exactly the same acquisition parameters. Representative images from the ChIP gels are shown. Quantitation of PCR products was performed using ImageLab software (Bio-Rad). The bound fraction values were calculated as a percentage of the input DNA used in the immunoprecipitation representing 100% and are shown as means of percentage bound from three independent experiments.
Histone Writers Atf2 and Dot1l Are Essential for CA-Driven stc2 Expression
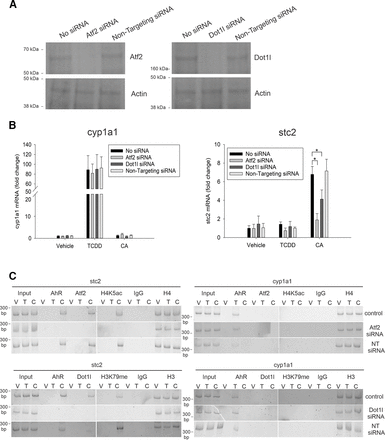
AML12 cells were transiently transfected with Atf2, Dot1l, and nontargeting (scrambled) siRNA oligonucleotides. Western blotting confirmed successful knockdown of Atf2 and Dot1l protein expression with RNA interference (Fig. 6A). Quantitative RT-PCR indicated that the loss of Atf2 and Dot1l significantly attenuated CA-induced stc2 expression, whereas the cyp1a1 message remained unaltered (Fig. 6B). ChIP studies revealed that silencing Atf2 obliterated AhR recruitment to the stc2 promoter and impeded H4 K5 acetylation (Fig. 6C). Similarly, suppressing Dot1l expression resulted in the loss of AhR and H4 K79me interaction at the stc2 promoter. Silencing histone modification writers had no effect on TCDD-driven AhR binding to the cyp1a1 promoter (Fig. 6C). Collectively, our data suggest that the CA-driven recruitment of chromatin regulators, Atf2, and Dot1l to the AhR-chromatin complex triggers histone epigenetic modifications, including histone H4 K5 acetylation and H3 K79 methylation exclusively at the stc2 promoter, which plausibly results in changes in chromatin structure and thereby induces stc2 expression.
Atf2 and Dot1l are required for the transcription regulation of stc2. (A) AML12 cells were transiently transfected with Atf2 and Dot1l siRNAs or nontargeting siRNA (NT siRNA). Twenty-four hours later, Western blotting on cell lysates was performed to monitor both Atf2 and Dot1l protein expression. Actin was used as a loading control. (B) AML12 cells, transiently transfected with Atf2, Dot1l, and nontargeting siRNA for 24 hours were treated with vehicle (DMSO), 6 nM TCDD, and 30 µM CA for 2 hours. Quantitative RT-PCR was performed to measure cyp1a1 and stc2 mRNA normalized to 18S rRNA. For statistical analysis, a mixed-effects multivariate ANOVA (MANOVA) model was used. After an overall significant F test from MANOVA model, the post hoc multiple-comparison tests were performed for the prespecified comparisons adjusted by Tukey procedure. *P < 0.05, and n = 3 independent batches of AML12 cells. (C) ChIP assays were performed on AML12 cells transiently transfected with targeted and NT siRNA and treated with vehicle, TCDD, and CA. PCR products were loaded onto two 5% polyacrylamide gels (represented by space), run, stained with SYBR Green, and imaged on Chemidoc MP imager (Bio-Rad) simultaneously with exactly identical acquisition parameters. n = 3 for stc2, and n = 2 for cyp1a1.
Role of Quaternary XRE Structure in Agonist-Specific Differential Gene Regulation
We examined whether the quaternary structure of the stc2 promoter encompasses complete epigenetic and structural information necessary for the agonist-specific recruitment of chromatin regulators, histone modifications, and AhR-mediated regulation of stc2. A modified AML12 cell line was constructed by replacing the 926-bp cyp1a1 promoter region containing 10 XREs (between −574 and −1500 bp from the transcription start site) with the stc2 promoter containing eight XREs (between −210 and −469 bp from the transcription start site, termed “stc2 XRE cassette”) using CRISPR/Cas9 editing (Fig. 7A). In the CRISPR/Cas9-edited AML12 cells, quantitative RT-PCR indicated upregulation of cyp1a1 gene expression in response to CA treatment (Fig. 7B). A marked reduction in cyp1a1 message upon TCDD treatment in edited cells was attributed to the lack of 10 XREs within the cyp1a1 promoter (Fig. 7B). ChIP studies performed in WT AML12 cells displayed direct binding of AhR to the cyp1a1 promoter upon TCDD treatment (Fig. 7C). In CRISPR/Cas9-edited AML12 cells—AhR, Atf2, and Dot1l—binding and interaction of H4 K5 acetylation and H3 K79 methylation to the “stc2 XRE cassette” within the cyp1a1 promoter was observed exclusively in response to CA treatment (Fig. 7C). These results strongly suggest that the agonist-specific AhR-mediated stc2 expression can be transferred with the stc2 promoter sequence in the context of chromatin architecture. Finally, this study confirmed that the quaternary DNA structure contains comprehensive epigenetic and higher-order chromatin conformational information necessary to elucidate agonist-specific differential gene regulation by AhR.
Agonist-specific AhR target-gene expression transfers with the gene-specific “XRE cassette” in the context of chromatin architecture. An edited AML12 cell line was constructed by replacing the 926-bp cyp1a1 promoter region containing 10 XREs (between −574 and −1500 bp from the transcription start site) with the stc2 promoter containing eight XREs (the 259-bp region termed “stc2 XRE cassette” – between −210 and −469 bp from the transcription start site) using CRISPR/Cas9 technology. (A) Illustration of cyp1a1 and stc2 promoter regions. Red and blue rectangles represent XREs (5′-GCGTG-3′) within cyp1a1 and stc2 promoters respectively. CRISPR/Cas9-edited AML12 cells, wherein the 259-bp stc2 XRE cassette is inserted by replacing cyp1a1 XREs within −574 and −1500 bp, is depicted. (B) WT (black bars) and CRISPR/Cas9-edited (gray bars) AML12 cells were treated with vehicle (DMSO), 6 nM TCDD, and 30 µM CA for 2 hours. Quantitative RT-PCR was performed to measure RNA expression of cyp1a1 and stc2 and normalized to 18S rRNA. For statistical analysis, a mixed-effects multivariate ANOVA (MANOVA) model was used. After overall significant F test from MANOVA model, the post hoc multiple-comparison tests were performed by Tukey procedure. *P < 0.05, n = 3 independent batches of AML12 cells. (C) Vehicle-, TCDD-, and CA-treated, WT and edited AML12 cells were subjected to chromatin immunoprecipitation using antibodies against AhR, H4 K5ac, Atf2, H3 K79me, Dot1l, and H3 (positive control). PCR products were fractionated and visualized on 5% polyacrylamide gels stained with SYBR Green. Samples were run on separate gels (represented by space), stained with SYBR Green, and imaged on Chemidoc MP imager (Bio-Rad) synchronously with exactly the same acquisition parameters (n = 3 independent batches of AML12 cells).
Discussion
Since its discovery in 1976, the aryl hydrocarbon receptor has been a pivotal transcription factor in both environmental toxicology and molecular pharmacology (Poland et al., 1976a,b). Apart from the identification of prototypical AhR ligands of anthropic origin, over the last 40 years, a number of natural, structurally diverse AhR ligands with varying binding affinities have been discovered (Denison and Nagy, 2003; Denison et al., 2011). Moreover, a recent identification of the endogenous AhR agonist CA presents a unique opportunity to study molecular mechanism underlying the CA- and TCDD-specific, mutually exclusive regulation of stc2 and cyp1a1 genes by AhR (Harper et al., 2013; Lowe et al., 2014; Joshi et al., 2015). Our previous studies using electrophoretic mobility shift assays showed AhR binding to radiolabeled oligonucleotide probes encompassing individual cyp1a1 and stc2 promoter XREs in response to both TCDD and CA treatments (Joshi et al., 2017). This observation suggested that the agonist-specific, AhR-mediated differential transcription regulation was at least in part dependent on the tertiary chromatin structure plausibly because of the distinct cofactor binding and specific epigenetic modifications. Very few studies have reported evidence of ligand-specific cofactor recruitment by AhR. In mammalian cell culture systems, ligand-selective interaction of AhR with several cofactors, including steroid receptor coactivator 1 (SRC1), steroid receptor coactivator 2 (SRC2), steroid receptor coactivator 3 (SRC3), thyroid hormone receptor-associated protein 220 (TRAP220), coactivator associated arginine methyl transferase 1 (CARM1), and peroxisome proliferator-activated receptor γ coactivator-1 (PGC1), was observed using two-hybrid assays (Zhang et al., 2008). Moreover, AhR agonists β-naphthoflavone and 3,3′-diindolylmethane displayed differential cofactor recruitment to the cyp1a1 promoter in Michigan Cancer Foundation-7 (MCF7) cells (Hestermann and Brown, 2003). Agonist-specific cofactor binding was also observed in other nuclear receptors, including glucocorticoid receptor (Monczor et al., 2019), androgen receptor (Muller et al., 2000), and epidermal growth factor receptor (Saeki et al., 2009). However, the findings presented here are unique because our results indicate CA-specific recruitment of distinct histone modification writers (Atf2 and Dot1l) to the AhR-chromatin complex, which results in specific epigenetic modifications (H4 K5ac and H3 K79me) at the stc2 promoter in vivo, which is responsible for regulating transcription of stc2.
Gene transcription is a highly orchestrated process that is tightly regulated by the local chromatin conformation (Chen and Li, 2010; Woodcock and Ghosh, 2010). The basic unit of chromatin, the nucleosome core, contains two copies of four types of histones (H2A, H2B, H3, and H4) that can be post-translationally altered into at least 80 known covalent modifications (Zhao and Garcia, 2015). These histone modifications produce “histone codes” that influence nucleosome compactness and chromatin organization, which results in activation or silencing of transcription (Kouzarides, 2007; Bannister and Kouzarides, 2011; Bartke and Kouzarides, 2011). Our previous mass spectrometry analysis performed on the liver nuclei isolated from TCDD- and CA-treated mice identified CA-specific H4 K5 acetylation (Joshi et al., 2017). Further studies confirmed that the H4 K5ac at the stc2 promoter was concomitant with the interaction of Mta2—a known chromatin modification “reader”—with the AhR (Joshi et al., 2017). To temporally catalog CA- and TCDD-specific histone modifications associated with AhR-bound chromatin, we performed xChIP-MS (Tian et al., 2012; Sowers et al., 2015; Tang et al., 2016). Parallel reaction monitoring, an ion monitoring technique based on high-resolution, high-precision mass spectrometry, was employed to simultaneously detect multiple histone modifications. Parallel reaction monitoring has a broad dynamic range, measures all transitions, and is more resistant to background noise than conventional selective reaction monitoring (Tang et al., 2014). xChIP-MS identified a myriad of AhR-bound stable and transient histone acetylations and methylations across genome, albeit we focused on CA- and TCDD-specific, mutually exclusive modifications. Histone H3 lysine 14 acetylation, H3 lysine 23 acetylation, and H3 lysine 27 dimethylation were uniquely observed at AhR-bound chromatin in response to TCDD treatment, whereas CA triggered stable association of H3 K79 methylation at the AhR-chromatin complex (Fig. 2). Surprisingly, we did not detect H4 K5 acetylation with high confidence possibly because of the limitation of signal detection by mass spectrometry, low abundance, or loss during sample preparation (Bensaddek and Lamond, 2016). Nevertheless, xChIP–Western blotting successfully confirmed presence of H4 K5ac at AhR-bound chromatin exclusively upon CA treatment (Fig. 3).
Modifications on histone H4, specifically lysine residues in N-terminal tail (lysine 5, 8, 12, and 16), are known to be involved in gene regulation and maintaining genome integrity (Turner et al., 1989; Shahbazian and Grunstein, 2007; Zheng et al., 2013). H4 K5 has largely been implicated in epigenetic priming (Park et al., 2013), bookmarking (Zhao et al., 2011), and transcription regulation (Borsos et al., 2015). Histone acetylation is controlled by families of nonredundant lysine acetyltransferase, which use acetyl CoA to form ε-N-acetyllysine on lysine residues of histone tails, neutralize the positive charge on histone lysines, decrease the DNA-histone interaction, open the chromatin structure, and facilitate recruitment of RNA polymerase II, resulting in transcription activation of target genes (Bartke and Kouzarides, 2011). Previous studies have identified and characterized Tip60, Hbo1 (Myst2), Cbp, p300, and Atf2 as known H4 K5 acetylation writers (Legube and Trouche, 2003). Moreover, several chromatin regulators are known to interact with AhR, including p300, Cbp, SRC1, transcriptional mediators/intermediary factor 2, p300/CBP Interacting Protein (Kobayashi et al., 1997; Beischlag et al., 2002). In this study, we reiterated interaction of histone acetylation “writers”—p300, Cbp and Tip60—to AhR irrespective of agonist specificity (Fig. 4). On the contrary, Atf2 was associated with AhR–stc2 promoter complex exclusively in response to CA treatment. Atf2 is a member of the activating transcription factor/cAMP-response element binding protein family of basic region leucine zipper proteins and a bona fide candidate reported to possess intrinsic lysine acetyltransferase activity (Nomura et al., 1993; Sheikh and Akhtar, 2019). Atf2 is known to interact with other transcription factors, bind to response elements on target genes, and stimulate distinct transcription programs. Association of Atf2 with AP1 is known to alter local DNA structure and initiate transcription, Atf2-Jun interaction mediates transcription of IFNβ, and Atf2 binding to βHLH-PAS family member HIF1α promotes its transcription activity (Falvo et al., 1995, 2000; Choi et al., 2009). Therefore, it is conceivable that CA-specific interaction of Atf2 with AhR can directly or by recruitment of additional lysine acetyltransferases acetylate histone H4 K5. The acetylated mark is then accessed by chromatin modification readers, such as previously identified Mta2 (Wu et al., 2013; Joshi et al., 2017) and Brd4, which employs the bromodomain to target the modified histone and regulate transcription (Shi and Vakoc, 2014).
This study also identified CA-specific stable association of H3 K79 methylation at the AhR-chromatin complex (Figs. 2 and 3). Histone H3 lysine 4, 36, and 79 methylation are the three histone H3 methylation marks known to be associated with an active form of chromatin (Hyun et al., 2017). ChIP-Chip arrays using H3 K79 methylation antibodies have revealed positive correlation of gene expression in mammalian cells with the recruitment of histone methyltransferase, Dot1l (Steger et al., 2008). Therefore, it is plausible that the CA-driven, AhR-mediated recruitment of histone modification “writers” Atf2 and Dot1l confer specific epigenetic marks including H4 K5ac and H3 K79me, remodel the chromatin structure, and provide access to the transcription machinery at the stc2 promoter. It is noteworthy that AhR and Atf2 recruitment to the stc2 promoter and H4 K5 acetylation correlates with the kinetics of the stc2 induction. However, binding of Dot1l and H3 K79 methylation occurs as early as 5 minutes after CA treatment (Figs. 1–5). Previous studies have noted kinetic discrepancies between binding of AhR, p300, SRC1, transcriptional mediators/intermediary factor 2, and p300/CBP Interacting Protein to the cyp1a1 promoter and cyp1a1 gene expression in response to different ligands (Hestermann and Brown, 2003). It is therefore feasible that additional transcription factors, coregulators, and signaling pathways, including retinoblastoma protein, E2 factor, RelA, nuclear factor ĸB, estrogen receptor, and nuclear factor-erythroid factor 2-related factor 2/musculoaponeurotic fibrosarcoma protein (Nrf2/Maf), might crosstalk with CA and CA-bound AhR and thereby influence stc2 regulation (Denison et al., 2011; Jackson et al., 2014). Further chromatin proteomic profiling (David et al., 2017) and ChIP-sequencing studies are warranted to address differences in the kinetics and its impact on the agonist-specific, AhR-mediated gene regulation.
A modified AML12 cell line was constructed by replacing known dioxin-responsive elements within the cyp1a1 promoter with the stc2 cassette containing eight XREs (Shen et al., 1991; Lusska et al., 1993). In the CRISPR/Cas9-edited AML12 cells, histone epigenetic modifications (H4 K5ac and H3 K79me) and chromatin modification “writers” (Atf2 and Dot1l) were transferred in conjunction with the quaternary structure of the stc2 promoter XREs, which resulted in a CA-dependent regulation of cyp1a1 expression. This reinforced the notion that the agonist-specific gene regulation by AhR is highly dependent on the ligand-specific cofactor recruitment and exclusive epigenetic signatures that influence chromatin architecture (Wajda et al., 2020). The diminished but persistent induction of cyp1a1 by TCDD in the edited cells is attributed to the functional XREs located beyond −1500 bp from the transcription start site, as acknowledged in the ChIP-sequencing studies (Fig. 7) (Nault et al., 2016). Our future studies beyond the scope of this manuscript will reveal genome-wide chromatin accessibility and nucleosome occupancy in response to CA versus TCDD treatments, determine presence of the CA-specific epigenetic signatures at other AhR target genes, identify molecular interactions of AhR with the “readers” and “erasers” of histone modifications, and will ultimately probe into the mechanics of chromatin remodeling in response to the AhR agonists. Collectively, these observations strongly demonstrate that the distinct cofactor binding and epigenetic modifications play critical role in the agonist-specific, AhR-mediated gene expression and highlight the complexities involved in the transcription regulation by AhR.
Acknowledgments
The authors thank Dr. Cornelis Elferink (University of Texas Medical Branch, Galveston) for the helpful discussions regarding xChIP-MS data analysis and CRISPR/Cas9 studies.
Authorship Contributions
Participated in research design: Joshi.
Conducted experiments: Patil, Tang, Rus, Joshi.
Contributed new reagents or analytic tools: Patil, Tang, Rus, Zhang, Joshi.
Performed data analysis: Patil, Tang, Zhang, Joshi.
Wrote or contributed to the writing of the manuscript: Patil, Tang, Zhang, Joshi.
Footnotes
- Received July 21, 2021.
- Accepted October 17, 2021.
This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK122028] and Presbyterian Health Foundation Harold Hamm Diabetes Center Seed Grant (to A.D.J.).
The authors report no conflict of interest.
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- AhR
- aryl hydrocarbon receptor
- AML12
- alpha mouse liver 12
- Arnt
- AhR nuclear translocator
- Atf2
- activating transcription factor 2
- CA
- cinnabarinic acid
- Cas9
- CRISPR-associated protein 9
- ChIP
- chromatin immunoprecipitation
- Dot1l
- disruptor of telomeric silencing 1-like histone lysine methyltransferase
- H3 K14ac
- histone H3 lysine 14 acetylation
- H3 K23ac
- histone H3 lysine 23 acetylation
- H3 K27dime
- histone H3 lysine 27 dimethylation
- H3 K79me
- H3 K79 methylation
- H4 K5ac
- H4 K5 acetylation
- HR
- homologous recombination
- MS/MS
- tandem mass spectrometry
- Mta2
- metastasis-associated protein 2
- PCR
- polymerase chain reaction
- RT-PCR
- reverse-transcription PCR
- rRNA
- ribosomal RNA
- siRNA
- small interfering RNA
- stc2
- stanniocalcin 2
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- WT
- wild type
- xChIP
- crosslinking ChIP
- xChIP-MS
- xChIP-coupled mass spectrometry
- XRE
- xenobiotic response element
- Copyright © 2021 by The American Society for Pharmacology and Experimental Therapeutics