Abstract
Chemokines such as stromal derived factor 1 and their G protein coupled receptors are well-known regulators of the development and functions of numerous tissues. C-X-C motif chemokine ligand 12 (CXCL12) has two receptors: C-X-C chemokine motif receptor 4 (CXCR4) and atypical chemokine receptor 3 (ACKR3). ACKR3 has been described as an atypical “biased” receptor because it does not appear to signal through G proteins and, instead, signals solely through the β-arrestin pathway. In support of this conclusion, we have shown that ACKR3 is unable to signal through any of the known mammalian Gα isoforms and have generated a comprehensive map of the Gα activation by CXCL12/CXCR4. We also synthesized a series of small molecule ligands which acted as selective agonists for ACKR3 as assessed by their ability to recruit β-arrestin to the receptor. Using select point mutations, we studied the molecular characteristics that determine the ability of small molecules to activate ACKR3 receptors, revealing a key role for the deeper binding pocket composed of residues in the transmembrane domains of ACKR3. The development of more selective ACKR3 ligands should allow us to better appreciate the unique roles of ACKR3 in the CXCL12/CXCR4/ACKR3-signaling axis and better understand the structural determinants for ACKR3 activation.
SIGNIFICANCE STATEMENT We are interested in the signaling produced by the G protein coupled receptor atypical chemokine receptor 3 (ACKR3), which signals atypically. In this study, novel selective ligands for ACKR3 were discovered and the site of interactions between these small molecules and ACKR3 was defined. This work will help to better understand the unique signaling roles of ACKR3.
Introduction
Chemokines are a family of small proteins that signal through the activation of G protein coupled receptors (GPCRs). The most evolutionarily ancient chemokine, C-X-C motif chemokine ligand 12 (CXCL12) or stromal derived factor 1 (SDF-1), signals through two receptors, C-X-C chemokine motif receptor 4 (CXCR4) and atypical chemokine receptor 3 (ACKR3), contributing to organogenesis, cancer metastasis, and HIV-1 infection, but the distinct biologic roles of the two receptors are not yet clear (Huising et al., 2003; Balabanian et al., 2005; Burns et al., 2006; Miller et al., 2008; Singh et al., 2013; Chatterjee et al., 2014; Wang et al., 2018). In addition to CXCL12, ACKR3 has an additional chemokine ligand, Interferon-inducible T-cell α chemoattractant (Burns et al., 2006). ACKR3, or C-X-C chemokine motif receptor 7 (CXCR7), is one of four atypical chemokine receptors, which are not thought to signal through G proteins but rather to signal solely through the β-arrestin pathway and are therefore described as “biased” receptors (Levoye et al., 2009; Rajagopal et al., 2010; Hoffmann et al., 2012; Bachelerie et al., 2013). However, there have been some reports of pertussis toxin-sensitive G protein signaling downstream of ACKR3 in primary rodent astrocytes (Odemis et al., 2012). Indeed, the ability of ACKR3 to couple to different G proteins has not been thoroughly investigated (Burns et al., 2006; Levoye et al., 2009; Rajagopal et al., 2010; Wang et al., 2018).
ACKR3 has a higher affinity for CXCL12 than CXCR4, so it is believed that one function of ACKR3 may be to sequester CXCL12 and modulate extracellular chemokine gradients as a way of regulating CXCR4 signaling (Crump, 1997; Balabanian et al., 2005; Sánchez-Alcañiz et al., 2011; Haege et al., 2012; Abe et al., 2018; Montpas et al., 2018). Early reports about ACKR3 noted its propensity to internalize after ligand binding and subsequently sequester its ligand from the extracellular space, giving rise to the suggestion that ACKR3 normally functions as a “decoy” or scavenger receptor (Balabanian et al., 2005; Naumann et al., 2010; Rajagopal et al., 2010). ACKR3 can also undergo internalization even when no ligand is present, and consequently constantly cycles between the plasma membrane and the cytoplasm (Naumann et al., 2010).
The role of CXCR4-signaling in cancer metastasis, HIV-1 entry into T-cells, and in the retention of hematopoietic stem cells in the bone marrow has made CXCR4 an important potential therapeutic target (Chatterjee et al., 2014; De Clercq, 2015). A small molecule antagonist for CXCR4, AMD3100 (Plerixafor) has found important clinical use in hematopoietic stem cell transplantation (De Clercq, 2015; Scala, 2015). The pharmacology of CXCR4 has been studied extensively and many small molecule ligands for CXCR4 have been described in the literature (Mishra et al., 2016; Peng et al., 2018).
In contrast to CXCR4, the pharmacology and biologic functions of ACKR3 are less well understood, although ACKR3 has been suggested as having a role in cancer pathogenesis where its expression is upregulated in several instances (Singh et al., 2013; Wang et al., 2018). Soon after ACKR3 was discovered to be a second receptor for CXCL12, the development of a series of ligands for the receptor was reported in the literature (Burns et al., 2006; Luker et al., 2009; Zabel et al., 2009). These original compounds from ChemoCentryx were shown to recruit β-arrestin to ACKR3. Although, interestingly, they are often referred to as ACKR3 “antagonists” when used in vivo, the reason for this disparity is unclear (Burns et al., 2006; Hartmann et al., 2008; Zabel et al., 2009; Rajagopal et al., 2010; Chen et al., 2015). Since then, other compounds have been shown to act as ACKR3 ligands, including the agonist VUF11207, which was developed using the ChemoCentryx ligands as a scaffold, and PF-06827080 (Wijtmans et al., 2012; Menhaji-Klotz et al., 2018). Two recent papers reported what appear to be the first two bona fide ACKR3 antagonists free of any ability to recruit β-arrestin (Menhaji-Klotz et al., 2020; Richard-Bildstein et al., 2020; Pouzol et al., 2021). Interestingly, several CXCR4 ligands have been shown to act differentially on ACKR3: the CXCR4 antagonists AMD3100 and TC14012 have been shown to act as ACKR3 agonists at elevated concentrations (Kalatskaya et al., 2009; Gravel et al., 2010; Montpas et al., 2015).
Several groups have sought to better understand how endogenous, peptide, and small molecule ligands interact with ACKR3. Of the papers that have examined how synthetic ligands interact with ACKR3, two reports focused on the small molecule ligands CCX777 and CCX622, whereas one paper examined the cyclic peptide ligand TC14012 (Montpas et al., 2015; Gustavsson et al., 2017; Gustavsson et al., 2019). It was demonstrated that CCX777 interacted with ACKR3 by binding in the deeper orthosteric pocket and TC14012 relies on residues D179 and D275, found on extracellular loops (ECL) ECL2a and ECL3, respectively (Montpas et al., 2015; Gustavsson et al., 2017). Although this is interesting, knowledge of the precise structural requirements for activation and binding of ACKR3 is constrained by limited availability of selective receptor ligands that do not activate CXCR4.
We now report the generation of a new group of ligands for ACKR3, together with an analysis of the parameters that determine ligand binding and receptor activation, including a comprehensive analysis of the ability of CXCR4 and ACKR3 to couple to all mammalian isoforms of the Gα subunit. These data further illuminate our understanding of the unique signaling characteristics of ACKR3 and supplements our knowledge concerning the structural requirements for ACKR3 activation.
Materials and Methods
Generation of Small Molecules and Other Ligands
Compounds NUCC-54129, NUCC-200823, NUCC-176289 and other NUCC analogs were synthesized according to methods described in US Patent 10,435,375 and in (Iyamu et al., 2019a, 2019b). NUCC analogs were dissolved in 20% DMSO and 80% water. CXCL12 was purchased through R&D (350-NS) and CXCL11 through BioLegend (574906). Fifty µg of CXCL12 was resuspended in 500 µl of PBS with 0.1% bovine serum albumin. PF-06827080, described as compound 18 in (Menhaji-Klotz et al., 2018), was acquired through Pfizer’s Compound Transfer Program. VUF11207 was purchased through Tocris (4780), AMD3100 through Sigma-Aldrich (A5602), and CCX771 and CCX733 through Chemocentryx. Quinpirole hydrocholoride, a D2 agonist, was purchased through Sigma (Q102).
Cell Lines
All cell lines were maintained as adherent cells in in Dulbecco’s Modified Eagle’s Medium (DMEM), 10% fetal bovine serum (FBS) and 1% penicillin streptomycin and kept in an incubator at 37°C with 5% CO2. High-titer, low-avidity cells are modified human embryonic kidney cells (HEK293) cells which express a TEV-protease fused to a β-arrestin 2 and a tTA-dependent luciferase reporter gene; these cells were used for the Tango assay (Barnea et al., 2008; Kroeze et al., 2015).
G Protein Assay
Experiments were performed as previously described (Masuho, et al., 2015a; Masuho et al., 2015b). An N terminally tagged myc-CXCR4 (98946, Addgene) and an N terminally tagged HA-ACKR3 were used (HG11535-NY, Sino Biologic). CXCL12 was resuspended as described above. Three independent experiments were conducted with HEK293 expressing masGRK3-Nluc reporter and the Gβ/γ subunits tagged with a fluorescent protein Venus. Cells were transfected with all of the G protein isoforms and either CXCR4, ACKR3, or no receptors. Bioluminescence resonance energy transfer (BRET) measurements were done on a microplate reader (POLARstar Omega, BMG Labtech) and measurements were taken over 1000 times every 0.1 seconds for all G proteins except Gαz, which were once every 0.22 seconds. Data for the traces are ΔBRET and the bar graphs show max ΔBRET. Normalized responses for CXCR4 were normalized to the largest response, in this case Gαi3. Max amplitude was calculated by taking the maximum ΔBRET value and kinetic rate constant (1/τ) of activation was calculated by fitting a single exponential curve to the traces with ClampFit software version 10.3. GraphPad Prism version 9 was used to run a one-way ANOVA for each individual Gα isoform and then a Tukey’s multiple comparisons test was used to test for differences between no receptors, CXCR4 and ACKR3.
β-arrestin Recruitment (Tango Assay)
On the first day of the three-day protocol, cells were plated on a 10 cm dish in DMEM with 10% FBS and 1% penicillin streptomycin. Twelve hours later, cells were transfected with the ACKR3-Tango (Addgene Plasmid #66265), CXCR4-Tango (no V2 tail) (Addgene Plasmid #66262), or D2-Tango (Addgene Plasmid #66269) (using Lipofectamine 3000 (cat # L3000001, Thermo Fisher Scientific). On the second day, 100,000 cells were transferred to each well of a poly-L-lysine (100 µg/ml)-coated 96 well-plate (Ref 655098, Greiner Bio-One) containing 200 µl of DMEM 10% FBS, 1% penicillin streptomycin medium. Twelve hours later, the appropriate volume of assay buffer (20 mM HEPES and 1x Hank’s balanced salt solution at pH 7.0) was added to make a final volume 300 µl, and appropriate volumes of drugs were added. The different experimental conditions were done in triplicate wells. As a control, three wells did not receive any compounds or CXCL12, “no ligand.” The next day, the medium and drugs were aspirated from the well and 100 µl of a 1:4 dilution of Bright-Glo (E2620, Promega) in PBS was added. Ten–15 minutes later, the luminescence levels were read three times with a 10-second delay between each reading using a Glomax 96 Microplate luminometer using the Glomax Bright-Glo protocol (Promega). GraphPad Prism version 9 was used to generate concentration-response sigmoidal curves and calculate log EC50 values using the “sigmoidal, dose log X” function. Data are shown normalized to the “no ligand,” baseline condition and represented as mean ± SD averaged across all three plate readings and across each well triplicate.
For comparison of logEC50 values of point mutations, if GraphPad labeled the sigmoidal curve as “ambiguous,” that logEC50 value was excluded. Any additional outliers were identified and excluded using the GraphPad Outlier calculator utilizing the Grubb’s test. A one-way ANOVA was run with Dunnett’s multiple comparisons tests to look for differences between wild-type (WT) and the mutations.
ACKR3 Mutagenesis
The point mutations at the D179, S103, K206, and D275 positions were generated by using a Quik Change II Mutagenesis Kit (200521, Agilent). Bacterial colonies were selected, scaled up, and then a miniprep (K210002, Invitrogen) was performed. The DNA collected from the miniprep was digested with ClaI (R097S, NEB) for four hours at 37°C and then run on an agarose gel. Samples with the correct band sizes were scaled up, a Maxiprep (D4202, Zymo Research) was performed, and then the DNA was sent to the NU Sequencing Core for Sanger sequencing.
Mutagenesis Primer Design
The following primers were used to generate point mutations in the ACKR3-Tango DNA using the Agilent kit as described above: D179N Forward: 5′ TGT GTT TCC TTG CCA AAC ACA TAC TAC CTG AAA ACC 3′ D179N Reverse: 5′GTT TTT CAG GTA GTA TGT GTT TGG CAA GGA AAC ACA 3′ D275N Forward: 5′ GTT GCA GTG CTG CTG AAT ATC TTT TCC ATA CTC 3′ D275N Reverse: 5′ GAG TAT GGA AAA GAT ATT CAG CAG CAC TGC AAC 3 ′K206D Forward: 5′ GAG CAT AGC ATC GAT GAG TGG CTC ATC GGC ATG 3′ K206D Reverse: 5′ CAT GCC GATG AGC CAC TCA TCG ATG CTA TGC TC 3′ S103D Forward: 5′ GTG TGG GTG GTG GAC CTG GTT CAA CAC AAC 3′ S103D Reverse: 5′ GTT GTG TTG AAC CAG GTC CAC CAC CCA CAC 3.’
The following primers were used for Sanger Sequencing through the Northwestern Sequencing Core for all mutants except S103D: Forward: 5′ TAC TTC ACC AAT ACC CCC TCC TCC 3′ and Reverse: 5′ GCA ATG CAC GAG AGA CAA ACA CTG 3′. For the S103D mutation, the following primers were used for Sanger Sequencing: Forward: 5′ CAG GCC AAG ACT ACA GGA TAC GAC 3′ and Reverse: 5′ GGA GGA GGG GGT ATT GGT GAA GTA 3′.
Immunohistochemistry
Cells from the second day of the Tango assay, expressing either WT or point-mutated versions of ACKR3-Tango, D2-Tango, or CXCR4-Tango receptors, were plated onto poly-L-lysine coated coverslips. Twenty-four hours later, coverslips were rinsed with PBS and fixed with 4% paraformaldehyde for 20 minutes at room temperature and stained for α-Flag using mouse primary antibody (F3165, Sigma) or α-ACKR3 (MAB42273, R&D Systems) at 1:500 overnight at 4°C, and a goat α-mouse Alexa 488 secondary antibody (Molecular Probes) was applied at 1:500 dilution for 1.5 hours at room temperature. The coverslips were mounted with Vectashield hard set mounting medium containing 4′, 6-diamidino-2-phenylindole (DAPI) (H-1500, Vector Laboratories). Cells were visualized using FVi10 Olympus confocal microscope using a 60x oil emersion objective and a 3x or 4x digital zoom with a 1μm step size. Images were processed using ImageJ (National Institutes of Health). The images shown were one optical slice of the confocal Z-stack.
cAMP Glo-Assay
Ten thousand cells were plated onto a 96-well, clear-bottomed plate (Ref 655098, Greiner Bio-One). The following day, the medium was aspirated and 20 µl of induction buffer (Krebbs Ringer Buffer with 10 µM forskolin, 100 µM Ro 20–1724, and 500 µM of IBMX) was added, along with the appropriate drug concentration. The cells were exposed to the drug for 45 minutes; afterward, 20 µl of cAMP-Glo Lysis (V1501, Promega) buffer was added for 15 minutes. Following lysis, 40 µl cAMP-Glo Reaction Buffer (2.5 µl of PKA was added for every 1ml of cAMP reaction buffer) for 20 minutes. Then, 80 µl of Kinase-Glo was added to each well. After a 10 minute incubation, the luminescence levels were read three times with a 10 second delay between each reading using a Glomax 96 well Microplate luminometer using the Glomax Kinase Glo protocol (Promega). GraphPad Prism version 9 was used to generate the concentration-response sigmoidal curves.
Binding Assay
HEK293 cells were plated onto 10 cm cell culture plates and, 24 hours later, transfected with HA-ACKR3 plasmid (HG11535-NY, Sino Biologic) or CXCR4-YFP plasmid using Lipofectamine 3000 (cat # L3000001, Thermo Fisher Scientific) (Toth et al., 2004). One day later, cells were collected and suspended in binding buffer (1x PBS/1.0% BSA/0.1% sodium azide). Reaction tubes were prepared on ice with cells, various concentrations of unlabeled CXCL12, or the small molecule compounds and the indicated concentration of CXCL12 AF647 (CAF-11, Almac Group). For competition-binding studies, 40 ng/ml CXCL12 AF647 was used and for Schild slope experiments, a range of CXCL12 AF647 was used from 20 ng/ml to 200 ng/ml or 0 ng/ml. All studies were performed at 4°C, competition-binding studies were 90 minutes for ACKR3 experiments, or 30 minutes for CXCR4 experiments, and saturation-binding experiments were done for 90 minutes. Cells were then spun down at 1500 rpm, washed twice with cold binding buffer, Pacific Blue DAPI (D1306, Life Technologies) was added, and the samples were then analyzed on BD LSR Fortessa 6 laser at Northwestern’s Robert H. Lurie Comprehensive Cancer Center-Flow Cytometry Core Facility. One-hundred percent binding was defined as mean fluorescence of transfected cells, plus CXCL12 AF647, and 0% binding was defined as untransfected HEK293 cells, plus CXCL12 AF647; these parameters were consistent with previously described methods (Szpakowska et al., 2018; Meyrath et al., 2020). Duplicate reaction tubes were prepared. Samples where the percentage of live cells, as evidenced by DAPI staining, was below 50% were not used for analysis. GraphPad Prism version 9 was used to generate concentration-response sigmoidal curves, calculate log IC50 values using the “sigmoidal, dose log X” function, calculate Hill Slope values, and to calculate one-way ANOVA with Tukey’s multiple comparison tests where described. GraphPad Prism version 9 was used to calculate Schild slope. For saturation-binding experiments, GraphPad Prism version 9 was used to calculate the Kd and Bmax using the “nonlinear regression (curve fit), one site total” function.
Results
G Protein Activation by ACKR3
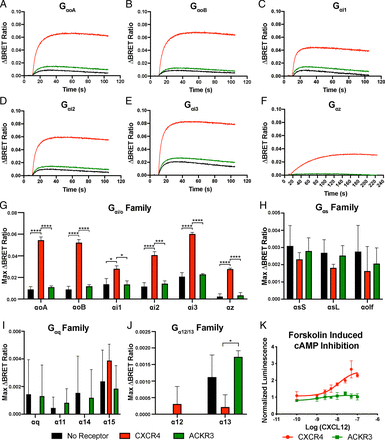
Given the disparities in the literature as to whether ACKR3 can effectively signal through G proteins, we examined the various mammalian isoforms of Gα proteins to comprehensively determine the spectrum of Gα subunits which could be activated by ACKR3 (Burns et al., 2006; Rajagopal et al., 2010; Odemis et al., 2012). We used a BRET assay in which HEK293 expressing masGRK3-NLuc reporter and the Gβ/γ subunits tagged with a fluorescent protein Venus. This assay detects G protein activation, resulting in the release of the Gβ/γ subunits by monitoring their interaction with masGRK3-NLuc in real time by increasing BRET ratios (Masuho et al., 2015b). Cells were transfected with either CXCR4, ACKR3, or no GPCR. Because chemokines are known to primarily signal through the Gαi/o family, we examined each of the mammalian isoforms belonging to that family—GαoA, GαoB, Gαi1, Gαi2, Gαi3, and Gαz—for their ability to be activated by CXCR4 and ACKR3 following application of 500 ng/ml CXCL12 (or SDF-1) (Fig. 1, A–F) (Wang et al., 2018). We also examined the ability of CXCR4 and ACKR3 to couple to members from the other families—Gαs, Gαq/11 and Gα12/13. The maximum ΔBRET for each group are shown in Fig. 1, G–J. Although ACKR3 was unable to couple to any of the isoforms we tested, CXCR4 was able to activate all of the members of the Gαi/o family with varying efficiency. A one-way ANOVA revealed significant differences, indicating productive coupling for all Gαi/o family members—GαoA, GαoB, Gαi2, Gαi3, Gαz (P < 0.0001), and Gαi1 (P = 0.0063)—but not for any members of the other major families (Fig. 1). A Tukey’s multiple comparison test showed a significant difference between cells expressing CXCR4 and cells with no receptors for all of the Gαi/o isoforms (* P = 0.015, **** P < 0.0001). CXCR4 was also able to activate Gαz, the only member of the Gαi/o family that is not sensitive to pertussis toxin (Ho & Wong, 2001). There were no significant differences between CXCR4 and the no receptor control for any members of the Gαs, Gαq/11 and Gα12/13 families, suggesting that CXCR4 can only signal through the Gαi/o family. Unlike CXCR4, there were no significant changes in BRET signal between cells without a receptor or cells with ACKR3 for any of the Gα isoforms, including the Gαi/o family.
Comparing CXCR4 and ACKR3′s ability to activate Gα isoforms. (A–F) Representative curves of ΔBRET ratios over time between cells transfected with either CXCR4 (red), ACKR3 (green), or no receptor (black), and the indicated G protein isoform from the Gαi/o family after the addition of 500 ng/ml of CXCL12 at 10 seconds. (G–J) The max ΔBRET values from the same experiment but with representatives from all major G protein families, Gαi/o (G), Gαs (H), Gαq (I), and Gα12/13 (J). (G–J) An ordinary one-way ANOVA followed by Tukey’s multiple comparisons for each Gα isoform showed no significant difference between the max value of ACKR3 and the no receptor control. A Tukey’s multiple comparisons test revealed significant differences between CXCR4 and the no receptor control for GαoA, GαoB, Gαi2, Gαi3, Gαz (**** P < 0.0001) and Gαi1 (* P = 0.0105). Data are shown mean ± SD across three independent experiments performed in triplicate. (K) A cAMP luciferase assay where a decrease in cAMP levels led to an increase in luminescence. Cells transfected with CXCR4 and treated with CXCL12 led to an increase in luminescence, whereas cells transfected with ACKR3 did not. Data are presented as mean ± SD performed in triplicate.
We then confirmed that the lack of ACKR3-mediated G protein activation was not due to insufficient expression of ACKR3 at the plasma membrane by performing saturation-binding experiments utilizing an Alexa 647 attached to the C terminus of CXCL12 (CXCL12 AF647) as our labeled ligand (Hatse et al., 2004; Janssens et al., 2016; Szpakowska et al., 2018; Meyrath et al., 2020) (Supplemental Fig. 1). These experiments were also performed on cells expressing CXCR4. Experiments were carried out at 4°C to inhibit receptor recycling. Under these conditions, ACKR3 was expressed at robust levels comparable to those of CXCR4 (Supplemental Fig. 1).
To further verify that the lack of G protein activation by ACKR3, we analyzed the ability of Gαi/o signaling to inhibit adenylate cyclase leading to a decrease in cAMP. When HEK293 cells were transfected with CXCR4, CXCL12 produced a concentration-dependent decrease in cAMP levels, resulting in an increase in luminescence. However, when cells were transfected with ACKR3, no comparable decrease in cAMP levels was observed (Fig. 1K).
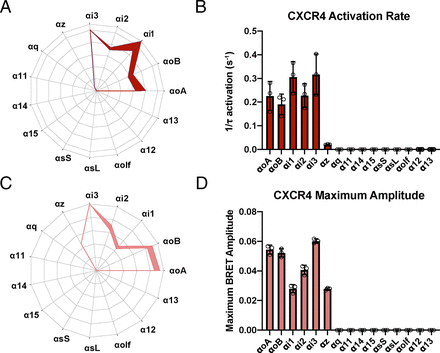
This data allowed us, for the first time, to make a comprehensive map of which Gα proteins were able to couple to CXCR4 and to examine the activation of G protein after CXCR4 stimulation more thoroughly (Fig. 2). Limited studies have been performed to examine the CXCR4/Gα interactions except for a few isolated reports that suggested CXCR4 may couple more efficiently to Gαi1 and Gαi2 compared with Gαi3 and Gαo, Gαq activated downstream of CXCR4, and Gα13 coupling in T-cells (Heuninck et al., 2019). As expected from the data shown in Fig. 1, the activation rate for all members of the Gαi/o were much higher than those observed with the other Gα families (Fig. 2, A and B). A one-way ANOVA of the kinetic activation rate showed significant differences (P < 0.0001). Tukey’s multiple comparisons showed many significant differences, both within the Gαi/o family and between Gαi/o and the other isoforms (Supplemental Table 1). We were also able to compare the maximum amplitude of the BRET signal generated by CXCR4 across all of the Gα isoforms tested. Not surprisingly, given the BRET responses observed in Fig. 1, all of the Gαi/o isoforms showed the largest amplitudes after CXCL12 stimulation of CXCR4 (Fig. 2, C and D). A one-way ANOVA of the kinetic activation rate showed significant differences (P < 0.0001). Tukey’s multiple comparisons revealed many significant differences in the amplitudes of the CXCR4-driven responses, both within the Gαi/o family and between Gαi/o and the other isoforms (Supplemental Table 1).
The Gα selectivity map of CXCR4. (A and C) A spiderweb showing either kinetic activation (A) or max amplitude (C) of CXCR4 coupling to different G protein isoforms after application of 500 ng/ml CXCL12. All responses were normalized to Gαi3, which had the largest response. (B) The activation rate (1/τ) of the different G proteins to CXCR4. An ordinary one-way ANOVA revealed a significant difference (P < 0.0001). (D) The maximum BRET amplitude from the different isoforms coupling to CXCR4. An ordinary one-way ANOVA revealed a significant difference (P < 0.0001). Tukey’s multiple comparisons tests from the one-way ANOVAs performed on B and D can be seen in Supplemental Table 1. Data are presented as mean ± SD across three independent experiments performed in triplicate.
Characterizing a Novel Group of Small Molecule Ligands for ACKR3
As described above, ACKR3 does not signal through G proteins, rather, its primary known mode of signaling is through the β-arrestin pathway (Balabanian et al., 2005; Rajagopal et al., 2010). To determine if novel small molecule ligands could lead to β-arrestin recruitment to ACKR3, we elected to use the Tango assay (Barnea et al., 2008; Kroeze et al., 2015). In this assay, an increase in β-arrestin recruitment leads to transcription of a luciferase reporter gene. We first tested ACKR3′s two endogenous ligands, the chemokines CXCL12 and CXCL11 (Supplemental Fig. 2). We observed that both of the endogenous ligands were able to induce concentration-dependent β-arrestin recruitment. Chemokine receptors, such as CXCR4 and ACKR3, are known to have high levels of constitutive activity where β-arrestin is recruited to the receptor without a ligand present (Naumann et al., 2010). This constitutive activity can be observed by examining the basal luminescence in the Tango assay. We observed that both CXCR4 and ACKR3 exhibited more luminescence in the absence of ligands in comparison with a non-chemokine GPCR, the dopamine D2 receptor, for example (Supplemental Fig. 3B).
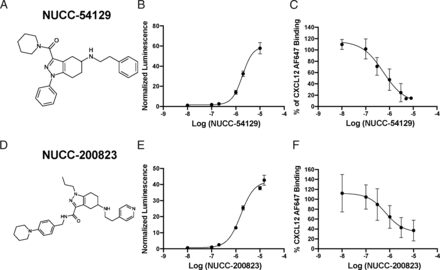
We recently described a series of novel heterocyclic compounds that activate CXCR4 signaling (Mishra et al., 2016). Because both CXCR4 and ACKR3 share a common ligand (CXCL12), we postulated that some members of this chemotype might also bind to ACKR3 and either activate or inhibit its function. To further explore the structure-activity relationships of this compound series for ACKR3, we synthesized a diverse library of approximately 250 tetrahydroindazole derivatives and tested them using the ACKR3 Tango assay described above. Each of the compounds retained the central tetrahydroindazole core and were diversified by varying the substituents off of the three attachment points. Examples of this set of compounds are provided in Supplemental Table 2. We found that many of the compounds tested had reproducible activity inducing β-arrestin recruitment to ACKR3. Although the majority of these compounds produced a concentration-dependent increase in β-arrestin signaling, 30 did not lead to an increase in luminescence at the highest concentrations tested. The two most potent compounds were NUCC-54129 and NUCC-200823, each with an EC50 in the low micromolar range (Fig. 3). The logEC50 values from the complete screen are shown in Supplemental Table 3. For comparison purposes, we also examined some of the most prominent small molecule ligands for ACKR3 curated from the literature including CCX771 and CCX733 (Burns et al., 2006; Luker et al., 2009; Zabel et al., 2009), VUF11207 (Wijtmans et al., 2012), AMD3100 (Kalatskaya et al., 2009), and PF-06827080 (Menhaji-Klotz et al., 2018) (Supplemental Table 4). All of these other ligands also led to β-arrestin recruitment in vitro, suggesting they have agonist activity (Supplemental Fig. 4).
Identification of two most potent small molecules in our library. (A and D) Structures of two of the most potent ligands in our library NUCC-54129 (A) and NUCC-200823 (D). (B and E) Representative concentration-response curves from the Tango assay with NUCC-54129 (B) and NUCC-200823 (E), with logEC50 values of −5.73 and −5.79, respectively. Data are presented as mean ± SD performed in triplicate. (C and F) Competition-binding assays with CXCL12 AF647 and either NUCC-54129 (C) or NUCC-200823 (F), with logIC50 values of −6.26 and −6.18 and Hill Slopes of −0.98 and −1.19, respectively. Data are presented as mean ± SD across 3 independent experiments performed in duplicate.
Importantly, we also tested the activity of our two most potent compounds, NUCC-54129 and NUCC-200823, along with NUCC-176289, a less potent ACKR3 agonist, at the CXCR4 receptor, the D2 dopamine receptor, and with cells that were not transfected with any GPCR (Supplemental Fig. 3). All three of these molecules produced concentration-dependent activation of ACKR3, but did not activate CXCR4 or D2 receptors, nor did they exhibit any activity in cells that were not transfected with a GPCR, indicating their selectivity for ACKR3 receptors (Supplemental Fig. 3, D–H).
A phenomenon of particular interest that we observed was that many of the small molecule ligands that we tested led to increases in maximum luminescence values well above those observed with the endogenous ligands CXCL12 and CXCL11. In some cases, small molecules produced activation as high as 100 times over the constitutive activity of the receptor, whereas the endogenous ligands led to luminescence levels around 10 times higher than baseline (Fig. 3; Supplemental Figs. 2 and 4).
To better understand how small molecule ligands interact with ACKR3, we employed the binding assay described above, utilizing CXCL12 AF647 as our labeled ligand (Hatse et al., 2004; Janssens et al., 2016; Szpakowska et al., 2018; Meyrath et al., 2020). Cells were treated with CXCL12 AF647 (40 ng/ml), either alone or in the presence of increasing concentrations of unlabeled CXCL12 (Supplemental Fig. 5A), or various concentrations of either NUCC-54129 or NUCC-200823 (Fig. 3, C and F). Both NUCC-54129 and NUCC-200823 were able to compete with CXCL12 AF647 for binding to ACKR3 with logIC50 values of −6.26 and −6.18, respectively. NUCC-54129 had a Hill Slope of −0.98 and NUCC-200823, −1.19. As previously reported, both PF-06827080 and VUF11207 were able to compete with CXCL12 for binding to ACKR3 (Supplemental Fig. 5, C and D). Interestingly, however, a Schild analysis of inhibition curves in the presence of 0.01 µM–10 µM NUCC-200823 and a range of concentrations of CXCL12 AF647 from 20 ng/ml to 200 ng/ml revealed a Schild Slope of 0.58, indicating that the mode of interaction between these novel small molecule ligands and the ACKR3 receptor may be complex.
We also tested whether NUCC-200841, which did not lead to β-arrestin recruitment in the Tango assay, could compete with CXCL12 for binding to ACKR3. NUCC-200841 was unable to block CXCL12 binding to ACKR3, which, combined with its inability to recruit β-arrestin, indicates that it is not active as a ligand for ACKR3 (Supplemental Fig. 5B). Importantly, we also showed that NUCC-54129 and NUCC-200823 were unable to compete with CXCL12 AF647 for binding to CXCR4, whereas CXCL12 was able to compete with CXCL12 AF647 for CXCR4 binding (Supplemental Fig. 5, E–G). Both NUCC-54129 and NUCC-200823 were unable to recruit β-arrestin to CXCR4 (Supplemental Fig. 3, E–H), nor compete with CXCL12 for binding to CXCR4, indicating that they are selective ligands for ACKR3 over CXCR4.
Generation of ACKR3 Point Mutants
Most chemokines follow a two-step, receptor-binding interaction with their receptors, first interacting with acidic residues on the N terminus and the extracellular loops (ECL) (chemokine recognition site 1) before the N terminus of the chemokine interacts with the transmembrane domains of the receptor (chemokine recognition site 2 [CRS2]) (Crump et al., 1997; Kleist et al., 2016). Although this model is perhaps oversimplified, it is interesting to note that one report has shown that the CXCL12/ACKR3 interactions do not appear to follow this canonical two-step binding mode (Kleist et al., 2016; Benredjem et al., 2017). Mutagenesis studies have also shown that the acidic residues in the N terminus are not essential for CXCL12 binding to ACKR3 (Benredjem et al., 2017). A different report has suggested that the N terminus helps extend the time that CXCL12 is bound to ACKR3, but that this interaction does not occur first and instead serves the purpose of preventing the CXCL12 from dissociating (Gustavsson et al., 2019). In contrast, CXCL11, ACKR3’s second endogenous ligand, does rely on the N terminus in a manner that follows the canonical chemokine two-step binding model (Benredjem et al., 2017; Gustavsson et al., 2019).
Several key amino acid residues were identified to be important for chemokine binding to their cognate receptors. The conserved residues D179 (4.60, Ballesteros-Weinstein numbering) and D275 (6.58) were found in ACKR3’s ECLs, ECL2a and ECL3, respectively (Montpas et al., 2015; Benredjem et al., 2017). D179 and D275 have been shown to be important for TC14021's, a CXCR4 antagonist that acts as a ACKR3 agonist, interaction with ACKR3 (Montpas et al., 2015). A homology model of ACKR3 also highlighted D179 and D275 as important residues for the receptor pharmacophore (Yoshikawa et al., 2013). Along with these two residues, K206 found in ECL2b is important for CXCL12 binding to ACKR3 (Benredjem et al., 2017). Evidence as to which residues may be involved in CRS2 have come from homology models and membrane-mimicking systems, which have identified some residues that are believed to be found in this deeper binding pocket (Gustavsson et al., 2017). Based on the crystal structure of CRS2 on CXCR4, S103 (2.63) and Q301 (7.39) have been identified as residues in the CRS2, orthosteric binding site, which are relevant for ACKR3 function (Benredjem et al., 2017).
We next examined whether different mutations would affect their ability to recruit β-arrestin to the receptor. We generated D179N, K206D, and D275N single amino acid substitutions in the ECLs on ACKR3-Tango receptor (Supplemental Fig. 6A). These amino acid residue switches were chosen to change the charge from acidic to uncharged (aspartic acid [D] to asparagine [N]) or basic to acidic (lysine [K] to aspartic acid [D]). Additionally, these substitutions were previously shown to impair CXCL12/ACKR3 interactions and, to a lesser extent, CXCL11/ACKR3 (Benredjem et al., 2017). We also chose to generate an S103D point mutation, found in the second transmembrane domain, because this residue corresponds to the D2.63 residue in CXCR4, which has been shown to be important for CXCL12 or small molecule ligand-binding to CXCR4 (Qin et al., 2015; Benredjem et al., 2017). When the S103 position was changed to reflect the residue found in the corresponding location in CXCR4, S103D, it was reported that there was decreased affinity for CXCL12 but no effect on β-arrestin recruitment (Benredjem et al., 2017).
It has already been shown that these mutations of ACKR3 do not affect folding or trafficking of the protein (Benredjem et al., 2017). To test the unlikely scenario that addition of the Tango elements to the C terminus of the receptor affected folding or trafficking, we used an antibody against the Flag tag of the Tango construct or an ACKR3 antibody to visualize expression of the mutated Tango constructs (Kroeze et al., 2015) (Supplemental Fig. 6B). As we expected, similar to the WT, all four of the point-mutated ACKR3 proteins had normal expression patterns and were observed on the plasma membrane and in the cytoplasm typical for a receptor such as ACKR3 with high constitutive activity (Naumann et al., 2010).
As mentioned above, the basal luminescence observed in the Tango assay reflects the high level of constitutive activity observed with ACKR3 (Supplemental Fig. 3B). Interestingly, the point mutations we generated had decreased constitutive activity to varying degrees, with S103D having the least and D275N having the most basal activity (Supplemental Fig. 7). A one-way ANOVA showed significant differences (P < 0.0001), and Dunnett’s multiple comparison showed a significant decrease in basal luminescence, evidence of constitutive β-arrestin activity, for the S103D (**** P < 0.0001), D179N (**** P < 0.0001), K206D (*** P = 0.0002) and D275N (* P = 0.0175) point mutations.
We next examined how small molecule ligands differed from the endogenous ligands in terms of interacting and initiating signaling with mutated ACKR3 receptors. We screened seven compounds against the four mutated versions of ACKR3—S103D, D179N, K206D, and D275N—along with unmutated WT receptors. The compounds chosen included two of the most potent novel compounds identified from our screen (Fig. 3) and several published ACKR3 agonists reported in the literature (Supplemental Fig. 4). Representative concentration-response curves from the Tango assay can be seen in Fig. 4 and the corresponding logEC50 values in Fig. 5 and Supplemental Table 5.
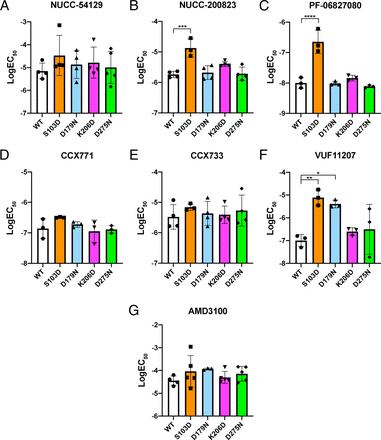
Concentration-response curves of small molecule ligand recruitment to mutated ACKR3. (A–G) Representative concentration-response curves from the Tango assay after application of various compounds with either WT or one of the four single amino acid-substituted versions of ACKR3 (S103D orange squares, D179N blue triangles, K206D pink inverted triangles, or D275N green diamonds). For each version of ACKR3, all values were normalized to a control where nothing was added for either WT or that specific mutated ACKR3. We tested various small molecules including two of our small molecules NUCC-54129 (A) and NUCC-200823 (B), an ACKR3 agonist from Pfizer PF-06827080 (C), two ChemoCentryx compounds, CCX771 (D) and CCX733 (D), VUF11207, which was modeled off of the ChemoCentryx compounds (F), and AMD3100 (G). Data are presented as mean ± SD performed in triplicate.
Changes in potency of small molecule ligand recruitment to mutated ACKR3. (A–G) LogEC50 values after application of a variety of small molecules to WT or one of the four single amino acid-substituted versions of ACKR3 (S103D orange squares, D179N blue triangles, K206D pink inverted triangles, or D275N green diamonds). LogEC50 values were compared using an ordinary one-way ANOVA, which revealed significant differences in EC50 values for NUCC-200823 (P = 0.0002) (B), PF-06827080 (P < 0.0001) (C), and VUF11207 (P = 0.0061) (F). Dunnett’s multiple comparisons test showed significant differences in logEC50 values for NUCC-200823 for WT and S103D (*** P = 0.0002) (B). Dunnett’s multiple comparisons test showed significant differences in logEC50 values for PF-06827080 for WT and S103D (**** P < 0.0001) (C). Dunnett’s multiple comparisons test revealed significant differences in logEC50 values for VUF11207 for WT and S103D (** P = 0.0055), and WT and D179N (* P = 0.0147) (F). Data are presented as mean ± SD from either three (C, D, F), four (B and E), or five (A and G) individual experiments, which were performed in triplicate.
In the case of NUCC-54129, all of the mutations led to an increase in efficacy, although there were no significant changes in logEC50 values (Figs. 4 and 5). NUCC-200823 showed an increase in efficacy with the D179N mutation. However, unlike NUCC-54129, NUCC-200823 showed a decrease in potency with the S103D mutation, with an average logEC50 of −4.87 ± 0.30 compared with −5.74 ± 0.10 for WT. A one-way ANOVA revealed a significant difference (P = 0.0002) and a Dunnett’s post-hoc test revealed a significant difference between WT and S103D (*** P = 0.0002); all other residues were not significant (Fig. 5).
PF-06827080 showed a dramatic decrease in potency with the S103D mutation, with an average logEC50 value of −7.65 ± 0.38 compared with −8.99 ± 0.17 for WT. A one-way ANOVA revealed a significant difference (P < 0.0001) and a Dunnett’s post-hoc test revealed a significant difference between WT and S103D (**** P < 0.0001); all other residues were not significant (Fig. 5). Interestingly, as was the case with NUCC-54129, PF-06827080 exhibited an increase in efficacy with both D179N and S103D (Fig. 4C).
CCX771 and CCX733 showed no significant differences in logEC50 values for any of the mutations when compared with WT. However, the concentration-response curves indicated that there was an increase in efficacy with the D179N mutation. The slight shift in the S103D concentration-response curves did not lead to any significant changes in the logEC50 values for S103D, nor any of the mutations for either CCX771 or CCX733 (Fig. 4, D and E, and Fig. 5).
VUF11207 is one of a series of compounds generated using the ChemoCentryx compounds as a template (Wijtmans et al., 2012). It therefore makes it especially interesting that VUF11207, CCX733, and CCX771 have different patterns of β-arrestin recruitment for the various mutations we generated. VUF11207 had an impaired ability to recruit β-arrestin with the D179N and S103D versions of ACKR3, with logEC50 values of −5.39 ± 0.17 and −5.11 ± 0.35, respectively, compared with WT −7.00 ± 0.27. A one-way ANOVA revealed a significant difference (P = 0.0051) and Dunnett’s post-hoc tests revealed a significant difference between WT and D179N (* P = 0.0147) and WT and S103D (** P = 0.0055) (Fig. 4F and Fig. 5).
AMD3100 is a very commonly used antagonist for CXCR4, but has also been shown to act as an agonist for ACKR3 (Kalatskaya et al., 2009; De Clercq, 2015). D179 and D275 are conserved with D171 and D262 on CXCR4, which are known to be important for AMD3100 binding (D4.60 and D6.58) (Labrosse et al., 1998; Montpas et al., 2015). Previous studies with AMD3100 on ACKR3 suggested that it could act allosterically because it could not block 125I-CXCL12 binding and had a small positive modulatory effect on β-arrestin recruitment when applied together with CXCL12 (Kalatskaya et al., 2009). When tested in the ACKR3-Tango assay with the various single amino acid substitutions, in contrast to other compounds, there was a decrease in potency with D179N, and none of the logEC50 values were significantly different (Fig. 4G and Fig. 5).
Discussion
We characterized a novel series of small molecule, nonpeptide ligands for ACKR3 receptors that can recruit β-arrestin to the receptor and compete with CXCL12 for binding to ACKR3 (Fig. 3). We then showed that the deeper binding pocket, particularly the S103 residue, was important for the agonist activity of some, but not all, of the small molecules we examined (Figs. 4 and 5).
In keeping with the majority of reports in the literature, our results showed that ACKR3 does not activate any mammalian isoforms of the α subunit of G proteins (Fig. 1). We also confirmed that this lack of signaling is not due to absence of ACKR3 surface expression (Supplemental Fig. 1). This result contrasts with a published report that ACKR3 can signal through Gαi/o proteins in astrocytes; however, it remains possible that the cell specific environment of astrocytes is required to enable G protein coupling (Odemis et al., 2012). However, our results were consistent with previous reports, utilizing both calcium imaging and BRET between ACKR3 and Gαi, in showing that ACKR3 was not able to couple with, nor signal through, G proteins (Burns et al., 2006; Levoye et al., 2009; Rajagopal et al., 2010). More work needs to be done to identify the molecular/structural elements that prevents ACKR3 and other atypical chemokine receptors from coupling with G proteins.
We screened over 250 novel small molecule ligands for their ability to recruit β-arrestin to ACKR3, identifying NUCC-54129 and NUCC-200823 as the most potent small molecules in our series (Fig. 3). These compounds have EC50 values in the low micromolar range and are selective for ACKR3 receptors over CXCR4. Both our novel ACKR3 ligands NUCC-54129 and NUCC-200823 competed effectively with CXCL12 for binding to ACKR3 (Fig. 3, C and F). The other ACKR3 ligands used in this study, except AMD3100, have also been reported to compete with CXCL12 for binding to ACKR3 (Supplemental Fig. 5) (Hartmann et al., 2008; Kalatskaya et al., 2009; Zabel et al., 2009; Wijtmans et al., 2012; Menhaji-Klotz et al., 2018). The addition of these small molecules to the limited pool of ligands for ACKR3 will allow us to better understand the structural determinants for ACKR3 activation.
Our results with CCX771, CCX733, and with other small molecule ligands once again confirm that, in vitro, these compounds act as agonists and lead to β-arrestin recruitment to ACKR3 (Supplemental Fig. 4 and Fig. 4) (Luker et al., 2009; Zabel et al., 2009). Interestingly, much of the literature has referred to both CCX771 and CCX733 compounds as ACKR3 “antagonists” (Burns et al., 2006; Rajagopal et al., 2010). It is possible that ACKR3 agonists lead to extensive receptor internalization in vivo and this might result in de facto antagonism, or that ACKR3 agonists produce receptor desensitization and loss of function (Huang et al., 2017; Lounsbury, 2020). The recent addition of two new bona fide nonsignaling ACKR3 antagonists should help us better understand how these ACKR3 agonists may lead to their observed effects in vivo (Menhaji-Klotz et al., 2020; Richard-Bildstein et al., 2020; Pouzol et al., 2021).
Strikingly, many of the small molecule ligands assessed in this study, including our two most potent compounds NUCC-54129 and NUCC-200823, led to luminescence responses as great as 100-fold above basal levels, whereas the endogenous ligands CXCL12 and CXCL11 led to luminescence levels around 10-fold (Fig. 3 and Supplemental Fig. 2). We speculate that this effect may be due to the ability of these small molecules to enter the cells and engage with ACKR3 receptors intracellularly, as opposed to the endogenous ligands which are confined to the extracellular space. Indeed, GPCRs that have been internalized can continue to signal from endosomes (Eichel et al., 2018; Naumann et al., 2010).
To examine the site of interaction of ACKR3 and small molecule ligands, we chose to focus on amino acid residues in the ECLs that have been shown to be important for CXCL12 binding, D179, D275 and K206, and S103 in the transmembrane domains region. We confirmed that the addition of the Tango elements did not impair the folding of the point-mutated ACKR3 receptors, which had previously been reported to fold and traffic properly (Supplemental Fig. 6) (Benredjem et al., 2017). Interestingly, we did observe some changes in the basal luminescence values, a measure of the amount of constitutive recruitment of β-arrestin to the mutated receptors without the addition of any ligand (Supplemental Fig. 7). Our results showed S103, situated in what is believed to be CRS2 for ACKR3, is a key residue for allowing several small molecule ligands to recruit β-arrestin to ACKR3, while not affecting the activity of endogenous chemokines (Figs. 4 and 5) (Benredjem et al., 2017). This is the case for small molecules that were generated in different medicinal chemistry pipelines such as NUCC-200823, PF-06827080, and VUF11207, suggesting that they are targeting a similar receptor binding site. A Schild analysis of NUCC-200823 revealed a slope of less than 1, so we cannot rule out that the binding mode of small molecules to CXCR7 may be mechanistically complex.
Although both VUF11207 and PF-06827080 exhibited decreased potency with the S103D mutation, the two most notable differences between the two compounds are that the D275N mutation has no effect with PF-06827080, whereas this change results in a greatly increased β-arrestin recruitment with VUF11207. In addition, the K206D mutation has little effect on PF-06827080, suggesting this compound does not significantly interact with this residue, whereas VUF11207 causes increased β-arrestin recruitment in the presence of the K206D mutant, indicating a more favorable interaction with an aspartic acid at residue K206 than a lysine. Because these two compounds have greatly different structures, it is not possible to clearly dissect their binding modes from this mutation data; however, it is clear that their interactions with residues D275 and K206 are different, although they may have similar interactions with residues D179 and S103 as mutations in these amino acids produce similar changes.
At this point, it is difficult to make an evaluation of the complete binding interactions between our new compounds and ACKR3 based on the mutation data, as we did not systemically survey a large number of compounds. However, it is clear that one or more of the substitutions of NUCC-54129 interact with S103, as modifying the three groups coming off of the central tetrahydroindazole core to produce compound NUCC-200823 result in a greatly attenuated ACKR3 β-arrestin recruitment. Future work to systematically test a large number of our new compounds in the presence of these point mutations, investigating one molecular change at a time, could allow for a detailed pharmacophore map to be generated and enhance our understanding of the precise binding mode of our compounds.
Overall, our results provide further insight into ACKR3 signaling and the addition of new small molecule ligands for ACKR3 (Fig. 3), which can help decipher the distinct biologic roles of CXCR4 and ACKR3. We showed that ACKR3 cannot activate any of the mammalian isoforms of Gα proteins and created a comprehensive map of CXCR4’s ability to couple to the Gαi/o family (Figs. 1 and 2). We also identified S103 as a crucial residue for small molecule interaction with the ACKR3 receptor (Figs. 4 and 5). Taken together, this work will help us further understand the significance of ACKR3 signaling and also ultimately allow us to better understand the distinct roles of ACKR3 in the CXCL12/CXCR4/ACKR3 signaling axis.
Acknowledgments
The authors would like to thank Bryan Roth and Wes Kroeze for assistance setting up the Tango assay, Matthias Majetschak for providing the ACKR3 Tango DNA, and Dr. Nevin Lambert for providing the Gβ/γ Venus construct. We would also like to thank Quinn Matusheck for her assistance with the ACKR3 S103D cloning. Additionally, we would like to thank Karen Ridge and Cara Gottardi for use of the luminometer and the Pfizer’s Compound Transfer Program.
Authorship Contributions
Participated in research design: Hopkins, Masuho, Martemyanov, Schiltz, Miller.
Conducted experiments: Hopkins, Masuho, Ren.
Contributed new reagents or analytic tools: Hopkins, Iyamu, Lv, Malik.
Performed data analysis: Hopkins, Masuho.
Wrote or contributed to the writing of the manuscript: Hopkins, Schiltz, Miller.
Footnotes
- Received April 3, 2021.
- Accepted May 31, 2022.
Research reported in this publication was supported by the National Institutes of Health National Cancer Institute [Grant R01-CA189074] (to G.E.S. and R.J.M.). This work was also supported by the Northwestern University-Flow Cytometry Core Facility supported by Cancer Center Support Grant [Grant NCI CA060553] and the Northwestern University NUSeq Core Facility.
The authors declare that they have no conflicts of interest with the contents of this article.
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- ACKR3
- atypical chemokine receptor 3
- BRET
- bioluminescence resonance energy transfer
- CRS2
- chemokine recognition site 2
- CXCL11
- C-X-C motif chemokine ligand 11
- CXCL12
- C-X-C motif chemokine ligand 12
- CXCR4
- C-X-C chemokine motif receptor 4
- CXCR7
- C-X-C chemokine motif receptor 7
- DAPI
- 4′,6-diamidino-2-phenylindole
- DMEM
- Dulbecco’s Modified Eagle’s Medium
- ECL
- extracellular loops
- GPCR
- G protein coupled receptors
- HEK293
- human embryonic kidney cells
- SDF-1
- stromal derived factor-1
- WT
- wild type
- Copyright © 2022 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.