Abstract
Voltage-gated KV1.3 channel has been reported to be a drug target for the treatment of autoimmune diseases, and specific inhibitors of Kv1.3 are potential therapeutic drugs for multiple diseases. The scorpions could produce various bioactive peptides that could inhibit KV1.3 channel. Here, we identified a new scorpion toxin polypeptide gene ImKTX58 from the venom gland cDNA library of the Chinese scorpion Isometrus maculatus. Sequence alignment revealed high similarities between ImKTX58 mature peptide and previously reported KV1.3 channel blockers—LmKTX10 and ImKTX88—suggesting that ImKTX58 peptide might also be a KV1.3 channel blocker. By using electrophysiological recordings, we showed that recombinant ImKTX58 prepared by genetic engineering technologies had a highly selective inhibiting effect on KV1.3 channel. Further alanine scanning mutagenesis and computer simulation identified four amino acid residues in ImKTX58 peptide as key binding sites to KV1.3 channel by forming hydrogen bonds, salt bonds, and hydrophobic interactions. Among these four residues, 28th lysine of the ImKTX58 mature peptide was found to be the most critical amino acid residue for blocking KV1.3 channel.
SIGNIFICANCE STATEMENT In this study, we discovered a scorpion toxin gene ImKTX58 that has not been reported before in Hainan Isometrus maculatus and successfully used the prokaryotic expression system to express and purify the polypeptides encoded by this gene. Electrophysiological experiments on ImKTX58 showed that ImKTX58 has a highly selective blocking effect on KV1.3 channel over Kv1.1, Kv1.2, Kv1.5, SK2, SK3, and BK channels. These findings provide a theoretical basis for designing highly effective KV1.3 blockers to treat autoimmune and other diseases.
Introduction
Autoimmune diseases are caused by excessive immune responses resulting from immune dysfunction. Epidemiologic studies showed that a large number of people in America suffer from autoimmune diseases, including multiple sclerosis, systemic lupus erythematosus, and type 1 diabetes (Miller et al., 2012; Wang et al., 2015). Although traditional autoimmune drugs such as steroids and cyclophosphamide can inhibit autoimmune responses and alleviate the symptoms caused by these responses (Teles et al., 2017; Matsubayashi et al., 2019), they lack selectivity and compromise normal protective immune responses, thereby increasing the chance of secondary infection, and severe side effects including nephrotoxicity, liver injury, and malignant tumors (Chandy et al., 2004; Wulff et al., 2019). Therefore, it is urgent to identify specific and therapeutic targets to selectively and effectively cure autoimmune diseases but maintain normal protective immune responses with minimal side effects.
Numerous studies indicate that the pathogenesis of autoimmune diseases such as multiple sclerosis, rheumatoid arthritis, and type 1 diabetes is mainly related to abnormal activation and proliferation of effector memory T (TEM) cells (Spanier et al., 2017; Falcao et al., 2018;Chemin et al., 2019). Normally, the body will not respond to self-antigens, but in autoimmune diseases or chronic infections, the TEM cells will be continuously activated by self-antigens and attack the peripheral tissues (Spanier et al., 2017; Chemin et al., 2019). Prior studies have shown that abnormal activation and proliferation of TEM cells was closely related to the high expression of KV1.3 channel (Matheu et al., 2008; Spanier et al., 2017). During immune responses, the expression of KV1.3 increased almost sixfold in activated TEM cells but stayed in a relatively low densities in naïve T cells or central memory T cells (Wulff et al., 2003; Zhao et al., 2015). Therefore, KV1.3 might be a specific drug target for autoimmune diseases because regulation of KV1.3 could selectively modulate the activation and proliferation of TEM cells without affecting the states of naïve T cells or central memory T cell (Perez-Verdaguer et al., 2016; Wang et al., 2019). Our recent study found that knocking out KV1.3 channel in Jurkat T cell with CRISPR/Cas9 technology inhibited almost half of Ca2+ influx and 90% interleukin-2 secretion upon activation (Shi et al., 2021). Other studies also showed that pharmacological inhibition and genetic ablation of KV1.3 function prevent the abnormal activation and proliferation of TEM cells in animal models of autoimmune diseases, while the protective immune responses to infective substances such as viruses and bacteria were maintained (Koni et al., 2003; Beeton et al., 2006; Matheu et al., 2008). These findings further point out that KV1.3 inhibitors are a potentially effective therapeutic drug for autoimmune diseases.
Metal ions, small organic molecules, and animal venom–derived polypeptides are three top candidates as potential KV1.3 inhibitors (Wulff et al., 2009; Zhao et al., 2015). Polypeptides have attracted attention because they have higher selectivity to ion channels and are part of the protein metabolism cycle bearing lower toxicities (Zhao et al., 2015; Wulff et al., 2019). Scorpion is an order of arachnids (Scorpiones) and its venom contains poplypeptides composed of 20–80 amino acid residues, which are important resources to discover KV1.3 channel inhibitors (Wulff et al., 2009; Wulff et al., 2019). In this study, we identified a new toxin polypeptide gene ImKTX58 from the cDNA library of Chinese Isometrus maculatus venom gland tissues. ImKTX58 gene-encoded peptide has high similarities with the known KV1.3 channel blockers-LmKTX10 and ImKTX88, which belong to the alpha-K+ channel toxin peptides (α-KTX) (Liu et al., 2009; Han et al., 2011). Whole-cell patch-clamp experiments showed that mature ImKTX58 peptide selectively and potently inhibited KV1.3 channels endogenously expressed in Jurkat T cells and heterologously expressed in HEK293T cells. Alanine scanning mutagenesis and computer simulation identified four amino acid residues (24th arginine, 28th lysine, 31st asparagine, and 37th tyrosine) in C-terminus of the ImKTX58 peptide that determine the potency of ImKTX58-induced inhibition of KV1.3 channel. These four residues interact with KV1.3 channel through hydrogen bonds, salt bonds, and hydrophobic interactions. Among these key residues, the 28th lysine was found to be the most critical residue required for the inhibition of KV1.3 channel. These structural and functional studies have established ImKTX58 peptides as effective and selective KV1.3 inhibitors, providing a foundation for developing therapeutic drug for KV1.3 channel-related autoimmune diseases.
Materials and Methods
cDNA Library Construction and Screening
I. maculatus scorpions were sampled from Hainan Province of China. Their species and sex were identified by an expert taxonomist. Twenty males and 20 females were used to build the venom gland library of the Chinese scorpion I. maculatus. Their glands were collected 2 days after electrical extraction of the venoms. Total RNA was extracted from 40 glands with Trizol reagent (Invitrogen/Thermo Fisher Scientific, Waltham, MA). Poly(A)-mRNA was purified by a PolyA Tract mRNA Isolation System (Promega, Madison, WI) and cDNA library was constructed according to the instruction of Superscript Plasmid System cDNA library construction kit (Gibco/BRL, Gaithersburg, MD). The cDNAs were cloned into pSPORT1 plasmids and Escherichia coli DH5α cells were transformed with the constructed plasmids. The positive clones of DH5α cells were randomly selected and sequenced to obtain the cDNA sequences of bioactive polypeptides from the venom gland of I. maculatus.
pGEX-4T-1-ImKTX58 Vector Construction and Toxin Site-Directed Mutagenesis
Primers for polymerase chain reaction were designed according to the cDNA sequence of mature ImKTX58 peptide. The forward primer was 5′-CTGGGATCCGATGACGATGACAAGCAGGTGCATACCAA-3′ with a BamHI restriction enzyme site (single underline) and an Enterokinase cleavage site (double underline). The reverse primer was 5′-CCGCTCGAGTCACCAATAGCAGGGCA-3′ with a XhoI restriction enzyme site (single underline). The template of polymerase chain reaction was total cDNA of I. maculatus venom gland tissues and the products were inserted into pGEX-4T-1 plasmid. A prokaryotic expression vector pGEX-4T-ImKTX58 was constructed and confirmed by sequencing. The QuickChange Site-Directed Mutagenesis Kit (Strategene, Bellingham, WA) was used to produce ImKTX58 mutated analogs based on the wild-type (WT) pGEX-4T-1-ImKTX58 plasmid. Universal pGEX primers were used to sequence all the plasmids. The plasmids with correct sequences were used to transform Escherichia coli Rosetta (DE3) cells to express ImKTX58 peptides and its mutated analogs.
Expression and Purification of ImKTX58 Peptides and its Mutated Analogs
E. coli Rosetta (DE3) cells were transformed with pGEX-4T-1-ImKTX58 plasmid or plasmids expressing its mutated analogs and proliferated at 37°C in LB medium with 100 mg/mL ampicillin. 0.5 mM Isopropyl b-D-thiogalactoside was added to the LB medium at 28°C for 4 hours to induce the fusion protein synthesis. The suspension was centrifuged and the supernatant was discarded. Glutathione-S-transferase (GST) wash buffer (pH 8.0, 50 mM Tris–HCl, 10 mM EDTA) was added to the precipitation to resuspend the cells and 1 mg/mL Lysozyme was applied to digest the cells for 30 minutes. After a brief sonication, the mixture was centrifuged at 10,000 × g for 15 minutes and the supernatant was collected. The fusion protein in supernatant was purified by GST affinity chromatography and concentrated with a 10 kDa molecular weight cutoff centrifugal concentrator (Millipore, Billerica, MA) (Yin et al., 2008; Bhuyan and Seal, 2015). High performance liquid chromatography (HPLC) was used to further purify the peptides and 230 nm wavelength UV radiation was applied to monitor the absorbance of elution at room temperature (22–25°C). The fusion protein was cleaved by Enterokinase (Biowisdom, Shanghai, China) for 8 hours at 37°C and the mixture was filtered (0.45 mm, Millex-HV, Millipore) and separated on a C18 column (10 mm×250 mm, 5 μm, Elite HPLC, Dalian, China). The elution buffer contained a linear gradient from 10 to 80% CH3CN with 0.1% TFA and the elution time length was 60 minutes with a constant flow rate of 5 mL/min. Peaks of the products were collected manually and lyophilized to powder, which was used for measuring the molecular weight by matrix assisted-laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
Mass Spectrometry and Circular Dichroism
The lyophilized peptides were purified by reversed-phase HPLC. The samples were mixed with 1 mL MALDI-matrix solution containing 10 mg/mL α-cyano-4-hydroxycinnamic acid, 0.1% trifluoroacetic acid and 45% acetonitrile, and spot on the MALDI target plate and air-dried at room temperature (25°C). Then the samples were tested on autoflex speed MALDI-TOF (Bruker Daltonics, Bremen, Germany). The mass of the peptides was measured with a positive ion linear mode and accelerating voltage was set as 20 kV. FlexControl software was applied for mass spectrometry analysis to obtain the mass-to-charge ratios ranging from 1000 to 8000 Da. The secondary structure of the polypeptides was determined by circular dichroism spectroscopy. The peptides were dissolved in 1 mL Milli-Q water at a concentration of 0.5 mg/mL. The circular dichroic spectrum was obtained by Chirascan (Applied Photophysics, Surrey, UK) at room temperature with the wavelength of 190–300 nm. The scanning speed is 50 nm/min, the response wavelength width is 1.0 nm, and the response time is 2 seconds. Each reading was repeated 3 times, and the results were displayed as the average residual molar ellipticity (θ).
Cell Culture and Potassium Channels Expression
Jurkat E6-1 T cells (TIB152, ATCC, Washington D.C.) and HEK293T cells (ACS4500, ATCC) were cultured in a humidified incubator at 37°C with 5% CO2 and maintained in Roswell Park Memorial Institute 1,640 basic (Cat. C11875500, Gibco, NY) or Dulbecco's modified Eagle's medium (Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (Life Technologies), 100 units/mL penicillin, and 100 μg/mL streptomycin, respectively. The coding sequences of mKV1.1, hKV1.2, and mKV1.3 were subcloned into the pIRES2-EGFP plasmid (Clontech, Mountain View, CA). HEK293T cells were transfected with the channel expressing plasmids for electrophysiological experiments. Lipofectamine 2000 (Invitrogen) was used for transfection of the plasmids.
Preparation of Dorsal Root Ganglion (DRG) Neurons
Experiment animals were bought from Wuhan Center for Disease Control and Prevention. All animal experiments were conducted according to the rules of the National Institutes of Health Guide for the Care and Use of Laboratory Animals and strictly following guidelines of the Institutional Animal Care and Use Committees. The Institutional Animal Care and Use Committees checked all protocols and approved this study. Female Kunming mice ages 3–4 weeks were anesthetized with 4% chloral hydrate (0.1mL/10g body weight) and killed. The whole spine was dissected out and put in ice-cold Hanks’ balanced salt solution. Extra muscles and tissues were discarded. The spine was cut along the middle line into two symmetrical parts with scissors. Dorsal root ganglions were picked under dissecting microscope. Fibers on DRGs were trimmed and only transparent ganglions were left. The round ganglions were lysised with 2 mg/mL Papain in Hanks’ balanced salt solution for 10 minutes under 37°C. During this time, the Eppendorf (Ep) tubes containing ganglions were shaken frequently. The Ep tubes were centrifuged under 4200 rpm for 4 minutes. The supernatant was discarded, and the precipitates were resuspended with 3.75 mg/mL collagenase and 3.75 mg/mL dispase for 10 minutes under 37°C. The Ep tubes were centrifuged under 4200 rpm for 4 minutes again. The supernatant was discarded and 1mL Dulbecco's modified Eagle's medium was added to stop enzyme digestions. DRG neurons were dissociated by trituration with a fire-polished glass Pasteur pipettes. Repeat centrifuging and suspending again and seeded the cells on coverslips preincubated with 10 µg/mL poly-D-lysine. Cells were cultured for 2 hours and extra culture medium (Neurobasal-A with 1 × B-27 supplement) was added. After more than 16 hours, DRG neurons were used for electrophysiology recordings.
Electrophysiological Recordings
Electrophysiological recordings were performed with an EPC9 amplifier (HEKA Elektronik, Lambrecht/Pfalz, Germany). Pipettes were pulled from borosilicate glass (BF 150-86-10; Sutter Instrument Company, Novato, CA). The resistances of the pipettes were 2-4 MΩ when filled with the internal solution. The internal and external solutions were prepared according to the previously described procedures (Yin et al., 2008). The internal solution for recording KV currents contained KCl 130 mM, MgCl2 1 mM, EGTA 5 mM, Na2ATP 3 mM, and HEPES 5 mM (pH 7.2 with KOH). The external solution contained KCl 4 mM, NaCl 137 mM, HEPES 10 mM, CaCl2 1.8 mM, MgCl2 1 mM, and D-Glucose 10 mM (pH 7.4 with NaOH). The internal pipette solution for recording NaV currents contained CsCl 145 mM, MgCl2 4 mM, EGTA 10 mM, Na2ATP 2 mM, HEPES 10 mM, and D-Glucose 10 mM (pH 7.3 with CsOH). The external solution contained NaCl 145 mM, KCl 2.5 mM, HEPES 10 mM, CaCl2 1.5 mM, MgCl2 1.2 mM, and D-Glucose 10 mM (pH 7.4 with NaOH). The internal solution for recording BK currents contained K-aspartate 145 mM, CaCl2 8.7 mM, MgCl2 2 mM, EGTA 10 mM, and HEPES 10 mM (pH 7.2 with KOH) to achieve an intracellular free Ca2+ concentration of 1 μM. The external solution contained Na-aspartate 130 mM, K-aspartate 30 mM, CaCl2 2 mM, MgCl2 1 mM, and HEPES 10 mM (pH 7.4 with NaOH). The internal solution for recording SK currents contained K-aspartate 140 mM, CaCl2 2 mM, hEDTA 5 mM, HEPES 10 mM, and Na2ATP 3 mM (pH 7.2 with KOH) to achieve an intracellular free Ca2+ concentration of 2.15 μM. The external solution contained Na-aspartate 140 mM, K-aspartate 5 mM, CaCl2 2 mM, MgCl2 1 mM, HEPES 10 mM, and D-glucose 10mM (pH 7.4 with NaOH). All electrophysiological experiments were conducted at room temperature (22–24°C).
For the current-clamp recording on DRG neurons, cells with rest membrane potential above −50 mV were discarded. Cells were clamped at the resting membrane potential and elicited with 0–1000 pA ramp current for 500 ms. The internal solution for current-clamp contained KCl 140 mM, NaCl 10 mM, MgCl2 1 mM, EGTA 1 mM, HEPES 10 mM, and MgATP 7.3 mM (pH 7.2 with KOH). The external solution for contained NaCl 154 mM, KCl 5.6 mM, CaCl2 2 mM, MgCl2 2 mM, HEPES10 mM, and D-glucose 1 mM (pH 7.4 with NaOH). After the cell state is stable, the action potentials of DRG neurons were recorded before and after drug administration. The number of action potentials, resting membrane potential and neuronal action potential peak were compared before and after the drug treatments.
Slice Preparation and Recordings
Brain slices were prepared as follows: mice (C57BL/6J; 21–28 days old; male) were anesthetized with isoflurane and decapitated rapidly, and 300 µm coronal slices of anterior piriform cortex was cut with Vibratome (Leica, Deutschland, Germany). Slices were cut in solution containing (in mM): 238 sucrose, 2.5 KCl, 10 glucose, 25 NaHCO3, 1.25 NaH2PO4, 0.5 CaCl2, 1.3 MgCl2, 2 MgSO4 (pH 7.2 with HCl), and transferred to the holding chamber containing artificial cerebrospinal fluid (ACSF). ACSF contained (in mM): 125 NaCl, 5 KCl, 11 glucose, 26 NaHCO3, 1.2 NaH2PO4, 2.6 CaCl2, and 1.3 MgCl2 (pH 7.2 with HCl). The opening resistances of the pipettes were 5–7 MΩ when filled with internal solution containing (in mM): 143.4 K-gluconate, 4 KCl, 10 HEPES, 0.3 EGTA, 10 phosphocretina-Na2, 4 MgATP, and 0.3 Na2GTP (pH 7.3 with KOH). Slices were incubated at 34°C for 45 minutes and recorded at room temperature. All the above processes were carried out in 5% CO2/95% O2 continuous bubbling.
The anterior piriform cortex (APC) is recognized according to its position relative to the lateral olfactory tract and the cell density of layer II. The pyramidal neurons of layer II in APC were identified by cell body morphology and positions, and recorded with multiclamp 700B amplifier (Molecular Devices, San Jose, CA). To record the Kv currents of pyramidal neurons, 1 µM TTX was added to block the sodium currents, the cell was clamped to −70 mV, and stimulated to +50 mV for 200 ms. For the current-clamp mode, currents were injected with 10 pA steps to trigger action potentials. The comparison between control and drug treatment was carried out under the same current stimulation.
Atomic Coordinates and Molecular Docking
The structure of ImKTX58 was modeled by Robetta structure prediction server (Supplemental Material 1). The homologous structure of the pore region of mouse KV1.3 channel (Supplemental Material 2) was built by using the human Kv1.3 channel (Protein Data Bank code: 7EJ1; Supplemental Material 3) as a template, because they have high sequence identity in the S5-P-S6 composing the pore region of the channels (Liu et al., 2021).
The interactions between ImKTX58 and mouse Kv1.3 channel were studied by molecular simulation using Autodock Vina program to generate the candidate complex structures of ImKTX58-KV1.3 (Chen et al., 2003). Possible hits were selected by clustering analysis and screening according to the experimental data. The candidate complexes were analyzed by further molecular dynamic (MD) simulation study.
The MD simulations were conducted with the Amber 11 program on a 128-CPU Dawning TC5000 cluster (Beijing, China). All the simulation steps were performed under the ff99 force field (Parm 99) (Yin et al., 2008). The unrestrained simulations and explicit solvent systems were applied to screen ImKTX58-KV1.3 complex. The SANDER module with periodic water box and the parameter of 1.5 ns equilibration and 15 ns unrestrained simulations was set in Amber11 program (Yi et al., 2007). The bilayer lipid membrane around the transmembrane helices of KV1.3 channel was not taken into consideration during the MD simulations since only the outer vestibule of KV1.3 channel was bound by ImKTX58 according to the mutagenesis studies. The force constant was gradually reduced from 5.0 (kcal/mol)/Å2 to 0.02 (kcal/mol)/Å2 in equilibration steps to restrain the heavy atoms in backbone. The temperature parameter for MD simulations was 300 K and the cutoff distance 10 Å.
To verify the quality and validity of the selected ImKTX58-KV1.3 complexes, the relative binding free energy ΔΔGbinding (differences in bond and free energy between mutant and WT) was calculated by molecular mechanics–generalized Born surface area module of AMBER 11. The main parameters to postprocess selected snapshots from the MD trajectories were as follows: The IGB value was 2 for activating the Onufriev’s GB parameters; the SURFTEN value was 0.0072 for computing the nonpolar solvation free energy with the LCPO method; the concentration of mobile counterions in solution was 0.1M for the SALTCON value; the EXTDIEL value of 80.0 was used as the dielectric constant for the solvent, and the INTDIEL value of 1.0 was set as the dielectric constant for the solution (Yi et al., 2007).
Statistics Analysis
GrapPad Prism (San Diego, CA) was used to analyze the data. All data are presented as mean ± SEM for n independent observations. Using IGOR (WaveMetrics, Lake Oswego, OR) software, concentration–response curves were fitted according to the following modified Hill equation:
 where I represent the peak current; [peptide] represents the concentration of toxin.
where I represent the peak current; [peptide] represents the concentration of toxin.
The IC50 was obtained by four-parametric nonlinear regression analysis constraining bottom to 0 and top to 1.
Results
Sequence Analysis of ImKTX58
To identify bioactive peptides from scorpion venom, we constructed a cDNA library of I. maculatus venom gland tissues and randomly selected and sequenced positive clones from the library to analyze the toxin transcriptome (Zhang et al., 2019). One positive clone was selected for further studies because it included a complete gene open reading frame (ORF) which could encode a toxin polypeptide. This peptide was named ImKTX58 (Im represents the abbreviation of the genus and species, KTX is the abbreviation of potassium channel toxin, and number 58 is the clone number in the cDNA library). For convenience of reference by other researchers, this peptide was also named kappa-Buthitoxin-Im1a according to the rational nomenclature introduced previously (King et al., 2008). The cDNA sequence of ImKTX58 is 404 bp in length. The 5′ and 3′ untranslated region of ImKTX58 were 129 and 95 bp, respectively (Fig. 1A). The 180 bp ORF could encode a peptide precursor consisting of 60 amino acid residues (Fig. 1A). A single AATAAA polyadenylation signal is 22 nt upstream of the poly(A) tail. The ImKTX58 precursor peptide contains a putative signal peptide of 22 residues predicted by SignalP V3.0 server (http://www.cbs.dtu.dk/services/SignalP/). A mature toxin of 38 residues with three pairs of disulfide bridges follows the signal peptide. Sequence alignment with other reported toxins revealed that ImKTX58 may also adopt the cysteine-stabilized–α/β scaffold structure of classic K+ channel blockers derived from scorpion venoms (Fig. 1B) (Liu et al., 2009; Han et al., 2011). The ImKTX58 shared 74% and 54% similarities with two other KV1.3 blockers—scorpion toxin LmKTX10 and ImKTX88—suggesting that ImKTX58 may also be able to block the KV1.3 channel.
Primary structure of ImKTX58. (A) Full-length ImKTX58 cDNA and protein sequences. The signal peptide is marked by single underline, the potential polyadenylation signal AATAAA is marked by double underline, cysteine residues is in red color, 5′ and 3′ untranslated regions in lowercase, and the numbers on the right represent the order of the amino acids. (B) Protein sequence alignment of ImKTX58 with its closest analogs.
Expression, Purification, and Characterization of ImKTX58 Peptide
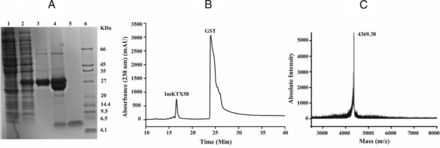
The GST affinity column was used to purify and molecular weight cutoff centrifugal concentrator (10 kDa, Millipore) to desalt the expressed GST-ImKTX58 fusion protein. The fusion protein was further cleaved by Enterokinase to separate GST protein and rImKTX58 peptides. The 30 kDa fusion protein was purified successfully and cleaved into the 26 kDa GST and the 4.4 kDa interest protein (Fig. 2A). The mixture after Enterokinase digestion was further separated by HPLC, which resulted in two peaks (Fig. 2B). The eluting component corresponding to recombinant ImKTX58 (rImKTX58) at about 17 minutes was collected and lyophilized for further analysis. The molecular weight of rImKTX58 was tested by MALDI-TOF-MS and the results showed the monoisotopic molecular mass of 4369.38 Da (Fig. 2C), which is consistent with the theoretical molecular weight of 4370.14 Da calculated with https://www.peptidesynthetics.co.uk/tools/.
Expression, purification, and identification of ImKTX58 peptide. (A) Tricine/SDS-PAGE analysis of the expressed ImKTX58 peptide. Lane 1, proteins extracted from noninduced E. coli Rosetta (DE3) cells; lane 2, proteins from E. coli Rosetta (DE3) cells transferred by pGEX-4T-1-ImKTX58 and induced by IPTG; lane 3, purified GST fusion protein after affinity chromatography analysis and desalting; lane 4, fusion protein cleaved by Enterokinase; lane 5, purified ImKTX58 by reversed phase HPLC; and lane 6, protein molecular weight marker. (B) Purification of ImKTX58 by HPLC on a C18 column. The peak at 17 minutes was ImKTX58 peptide, and the peak at 25 minutes was GST protein. (C) Mass spectrum of ImKTX58 peptide measured by MALDI-TOF-MS. The monoisotopic molecular mass of ImKTX58 measured by MALDI-TOF-MS is 4369.38 Da, and the calculated one is 4370.14 Da.
ImKTX58 Is a Selective KV1.3 Channel Blocker
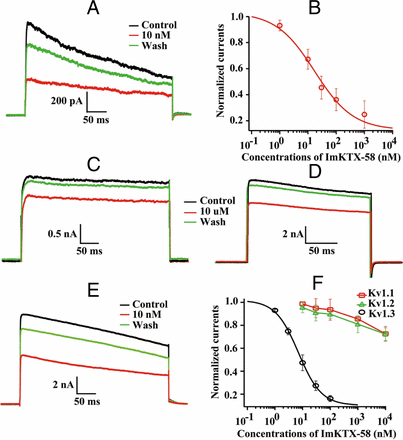
Sequence alignment showed that ImKTX58 polypeptide has high similarities with KV1.3 channel blockers LmKTX-10 and ImKTX-88 (Fig. 1B), suggesting that ImKTX58 may also inhibit KV1.3 channel (Liu et al., 2009; Han et al., 2011). We first examined whether ImKTX58 regulates the activation of KV1.3 channels endogenously expressed in human T cell line–Jurkat T cells. To avoid activation of the small conductance calcium-activated potassium channel 2 (SK2) channel in this cell, we used a Ca2+-free pipette solution. KV1.3-mediated currents were elicited by +50 mV depolarizing pulses for 400 ms from a −60 mV holding potential. Bath application of ImKTX58 reduced KV1.3 currents measured at the end of the depolarizing pulse (Fig. 3A). The IC50 value obtained by the fitness of concentration–response curve is around 39.41 ± 11.4 nM (n = 7) (Fig. 3B). The inhibitory effect of ImKTX58 was partially reversed after washout (Fig. 3A).
Modulation of endogenously and heterologously expressed voltage-gated K+ channels by ImKTX58. (A) Representative traces of KV1.3 current under Control (black), 10 nM ImKTX58 (red), and Wash (green) in a Jurkat T cell. (B) Concentration–response curve of KV1.3 current with ImKTX58 treatment. Current amplitudes were normalized to the Control group and fitted by Hill equation. (C–E) Representative current traces of KV1.1, KV1.2, and KV1.3 channels expressed in HEK293T cells in the absence (Control) or presence of ImKTX58. (C) 10 μM ImKTX58 on KV1.1, (D) 10 μM ImKTX58 on KV1.2, and (E) 10 nM ImKTX58 on KV1.3. (F) Concentration–response curves of ImKTX58-induced inhibition on KV1.1, KV1.2, and KV1.3 currents. KV currents were elicited by +50 mV, 400 ms depolarizing pulse every 30 seconds from the holding potential of −60 mV. The y-axis is the normalized current amplitudes, and the x-axis is the concentration of ImKTX58 peptide. Data are represented as mean ± SEM of at least five batches.
We next asked whether ImKTX58 also inhibits KV1.3 channels heterologously expressed in HEK293T cells. Consistent with the results from the Jurkat T cells, the peak amplitude of WT mKv1.3-mediated currents could be reduced by ImKTX58 and the inhibition rates were concentration-dependent (Fig. 3, E and F). The steady-state current at the end of the depolarizing pulse was measured and ImKTX58 could markedly decreased the amplitudes of the currents with an IC50 value of 10.42 ± 1.46 nM (n = 7) (Fig. 3F).
Mammalian KV1.1 and KV1.2 channels are highly homologous to KV1.3 and their structure homology is the main factor affecting the selectivity of KV1.3 channel blockers (Kim and Nimigean, 2016), so we determined whether ImKTX58 also inhibits heterologously expressed KV1.1 and KV1.2 channels in HEK293T cells. Surprisingly, application of 10 μM ImKTX58—1,000-fold higher concentration than that applied to KV1.3 channel—only reduced about 27% and 28% of KV1.1 (Fig. 3C) and KV1.2 (Fig. 3D) currents, respectively, suggesting that ImKTX58 has a higher selectivity for KV1.3 over KV1.1 and KV1.2.
To further confirm the selective inhibition on Kv1.3 channel, we also applied ImKTX58 on voltage-gated sodium (Nav) channel heterologously expressed in HEK293T cells. Our results showed that 10 μM ImKTX58 could only change the Nav1.4 currents to 103.93 ± 0.81% (n = 3), Nav1.5 currents to 94.43 ± 4.25% (n = 4), Nav1.7 currents to 103.83 ± 4.95% (n = 3) of the control groups (Fig. 4, A–C). Besides Kv1.1 and Kv1.2, the effects of ImKTX58 on calcium-activated potassium channel and Kv1.5 were also tested. The inhibition rate of 10 μM ImKTX58 on BK, SK2, SK3, and Kv1.5 were separately 10.42 ± 0.93% (n = 3), 3.93 ± 0.81% (n = 3), 14.04 ± 2.68% (n = 3), and 12.91 ± 3.64% (n = 4) (Fig. 4, D–G). These results suggest that ImKTX58 could be developed into KV1.3 channel blocker because it is highly selective to Kv1.3.
Effects of ImKTX58 on Nav and calcium-activated potassium channels exogenously expressed in HEK293T cells. (A–C) Example current traces of of NaV1.4, NaV1.5, and NaV1.7 channels expressed in HEK293T cells in the absence (black), presence of 10 μM ImKTX58 (red) and wash with external solution (blue); (D–F) Example current traces of BK, SK2, and SK3 channels expressed in HEK293T cells under Control, presence of 10 μM ImKTX58 and wash with external solution; (G) Example current traces of Kv1.5 channel expressed in HEK293T cells in the absence, presence of 10 μM ImKTX58 and wash with external solution. Nav currents were elicited from −80 mV to −20 mV for 100 ms. For BK currents recording, the membrane potentials were clamped to −100 mV for 50 ms, followed by a 400 ms voltage ramp from −100 to +100 mV and the holding potential was −60 mV during the 5-second sweep interval. For SK2 currents recording, the membrane potentials were clamped to −100 mV for 50 ms, followed by a 400 ms voltage ramp from −100 to +100 mV, then clamped to +100 mV for 50 ms and held at 0 mV during the 5-second interval. For SK3 recording, the membrane potentials were clamped to −100 mV for 50 ms, followed by a 400 ms voltage ramp from −100 mV to +100 mV and held at 0 mV during the 5-second interval.
The Effects of ImKTX58 on Action Potentials
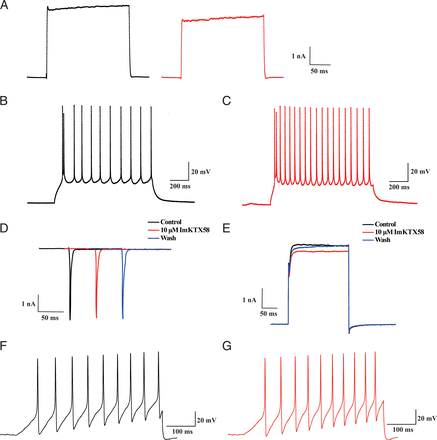
We have tested the effects of ImKTX58 on Kv channels in Jurkat T cells that mainly express Kv1.3 and SK2 (Valle-Reyes et al., 2018). Our results showed that ImKTX58 could inhibit the Kv currents in this kind of T cell line. Besides immune cells, Kv1.3 is also expressed in the nervous system and plays important roles in regulating the excitability of neurons. It is of interest to test the effects of ImKTX58 on excitable cells. We applied ImKTX58 on layer II pyramidal cells in anterior piriform cortex (APC). This group of neurons has been reported to express Kv1.3 channel (Al Koborssy et al., 2019). Our results showed that 10 μM ImKTX58 could significantly inhibit the Kv currents in these pyramidal cells (Fig. 5A). The inhibition rate of ImKTX58 on Kv currents in pyramidal cells was 27.27 ± 1.66% (n = 3). The frequency of the action potential was increased 2.42 ± 0.48-fold by ImKTX58 treatment, while the peak amplitude of the action potential was slightly decreased by 7.39 ± 2.45% (n = 3) (Fig. 5, B and C). And the rest membrane potential was slightly elevated by 5.05 ± 1.61% (n = 3). These results indicate that ImKTX58 could increase the excitability of pyramidal neurons in APC.
Effects of ImKTX58 on Kv currents and action potentials of neuronal cells. (A) Example current traces of endogenous Kv currents in pyramidal cells before (left, black) and after (red, right) 10 μM ImKTX58 administration, the ImKTX58 treatment time length was 15 minutes. (B, C) Representative action potential traces of pyramidal cells before (B) and after (C) 10 μM ImKTX58 administration for 15 minutes. (D) Example current traces of endogenous Nav currents in DRG neurons under Control, 10 μM ImKTX58 and Wash treatments. (E) Example current traces of endogenous Kv currents in DRG neurons under Control, 10 μM ImKTX58 and Wash treatments. (F, G) Representative action potential traces of DRG neurons before (F) and after (G) 10 μM ImKTX58 administration.
To compare the effects of ImKTX58 on peripheral neurons, it was also applied on cultured DRG neurons. Nav (n = 3) and Kv (n = 4) currents were not significantly changed by ImKTX58 (Fig. 5, D and E). And the frequency or peak amplitude of action potential were not significantly affected (Fig. 5, F and G). This result is consistent with the previous reports that the DRG neurons mainly express Kv1.1 and Kv1.2 channels and the expression of Kv1.3 was much lower than these two channels (Yang et al., 2004). Because10 μM ImKTX58 has little effect on Kv1.1 and Kv1.2, the Kv currents and action potentials in DRG neurons were not significantly affected. These studies on action potentials of neuronal cells further confirmed the specificity of ImKTX58 inhibition on Kv1.3.
The Structural Basis of ImKTX58 Regulation of Kv1.3 Channel
At the concentration of 10 nM, ImKTX58 inhibited about half of KV1.3 channel current, in contrast, previously reported KV1.3 channel inhibitors like Orthochirus scrobiculosus toxin-1 and Autoimmune Drug from Wenxin group (ADWX-1) could inhibit KV1.3 current in picomolar concentrations (Yin et al., 2008; Bhuyan and Seal, 2015). Therefore, we explored the molecular mechanisms underlying the inhibitory effect of ImKTX58 on KV1.3 channel to optimize the ImKTX58 structure and improve its blocking efficiency on KV1.3 channel. In general, the scorpion toxin polypeptides that modulate the ion channels share a conserved structure, comprising an α-helix at the N-terminus and 2–3 reverse antiparallel β-strands at the C-terminus and stabilized by 2-4 disulfide bonds (Giangiacomo et al., 2004). Amino acid residues at the C-terminus of the scorpion toxin polypeptides are critically involved in the recognition of the pore region of KV channels and mediate the channel blocking effects through electrostatic or hydrogen bonding and van der Waals's forces (Giangiacomo et al., 2004; Liu et al., 2009; Han et al., 2011). To identify the key residues involved in the interaction between ImKTX58 and KV1.3 channels, seven amino acid residues at C-terminus were individually mutated to alanine. The circular dichroism (CD) spectra of seven mutants showed no significant changes compared with the WT ImKTX58 peptide (Fig. 6I), confirming that ImKTX58 and its mutant analogs all adopted the same conformation.
Effects of ImKTX58 and its mutants on KV1.3 channel. (A–H) Representative KV1.3 current traces blocked by ImKTX58 and its mutants: (A) 10 nM ImKTX58, (B) 1 μM ImKTX58-R24A, (C) 100 nM ImKTX58-H26A, (D) 10 μM ImKTX58-K28A, (E) 100 nM ImKTX58-M30A, (F) 3 μM ImKTX58-N31A, (G) 300 nM ImKTX58-K32A, and (H) 1 μM ImKTX58-Y37A. (I) Circular dichroism spectra of ImKTX58 and its mutants. (J) Concentration-dependent inhibition of KV1.3 channels by ImKTX58 and its mutants.
The blocking effects of these ImKTX58 mutants on KV1.3 channel were then tested with electrophysiological experiments. All mutants showed decreased inhibition on KV1.3 current compared with the WT ImKTX58 peptide (Fig. 6, A–H, 6J and Table 1). The 24th arginine (Arg24), 28th lysine (Lys28), 31st asparagine (Asn31), and 37th tyrosine (Try37) mutants exhibited the most significant decreases in ImKTX58-induced inhibition on KV1.3 channel for over 91-, 960-, 155- and 48-fold (Fig. 6J), suggesting Arg24, Lys28, Asn31, and Try37 were the primary functional residues responsible for ImKTX58 interaction with the KV1.3 channel.
IC50 values for inhibition by ImKTX58 and its mutants on KV1.3
Simulation of the Interaction Between ImKTX58 and mKV1.3 Channel
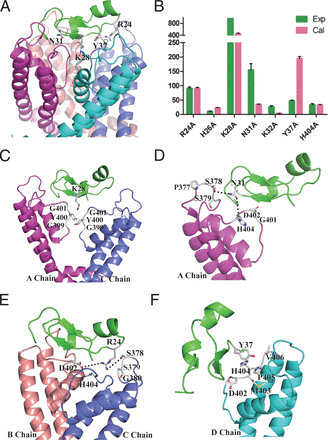
To better elucidate the interaction between ImKTX58 and KV1.3 channel, the stable structure of ImKTX58-KV1.3 complex was obtained through homology modeling, molecular docking and molecular dynamics simulation. Since our electrophysiological results identified Lys28, Asn31, Arg24, and Try37 as the key players between ImKTX58 and KV1.3 interaction, the ImKTX58-KV1.3 complex with these four amino acids located in the binding surface was screened first (Fig. 7A). To further verify the rationality of the simulated structure, the calculated ΔΔGbinding through computer alanine scanning technique and IC50 values of all mutants from electrophysiological results were normalized to WT ImKTX58 and compared with each other. Both the calculated and the experimental data showed that the pore blocking amino acid Lys(K)28 has a higher affinity to KV1.3 channel than the other amino acids (Fig. 7B). Moreover, the binding activities of Asn(N)31 and Tyr(Y)37 and Arg(R)24 are higher than that of His(H)26 and K32. It is worth noting that the H404 amino acid residue of KV1.3 plays an important role in the interaction with ImKTX58, which is consistent with the electrophysiological findings that the IC50 of ImKTX58 on Kv1.3-H404A was 351.98 ± 41.4 nM, about 33.78 times higher than that of WT KV1.3 (Supplemental Fig. 1).
Structural view of ImKTX58-KV1.3 complex and comparison of calculated and experimental results among ImKTX58 mutants. (A) An overview of the final model of the ImKTX58-KV1.3 complex showing the critical Lys28, Asn31, Arg24 and Tyr37 residues of ImKTX58. (B) The comparison between experimental and calculated data on the binding affinity toward KV1.3 channel for the seven Alanine mutants. The calculated results are normalized values of ΔΔGbinding, which was obtained from 1.5 ns restrained MD simulations and 15 ns unrestrained MD simulations, whereas experimental results are IC50 (Mutant)/IC50 (WT). (C–F) Interaction details of Lys28 (C), Asn31 (D), Arg24 (E), and Tyr37 (F) residues in ImKTX58 with KV1.3, respectively.
The IC50 of ImKTX58-K28A for KV1.3 channel was higher than 10 μM, almost 1,000-fold compared with the IC50 of WT ImKTX58. As shown in the complex structure of KTXIm58-KV1.3 in Fig. 5C, Lys28 in ImKTX58 was surrounded by the conserved “GYG” motif in the selectivity filter of KV1.3 channel within a range of 4 Å. This kind of structure is conducive to forming a strong interaction force, suggesting a high affinity to KV1.3 channel by Lys28 in WT ImKTX58. The importance of the Lys28 is also reflected by the fact that other KV channel toxin inhibitors usually insert a lysine side chain into the central pore of the channel and form a structure similar to a cork in bottle (Banerjee et al., 2013). Besides Lys28, the simulation results showed that N31 of ImKTX58 interacts with P377, S378, and S379 in the A chain and G401, D402, and H404 in the B chain of KV1.3 channel within a contact distance of 4Å (Fig. 7D). In addition, R24 of ImKTX58 also plays an important role in the interaction between ImKTX58 and KV1.3 channel as it is surrounded by the polar “groove” that is formed by D402 in B chain and S378 in C chain of KV1.3 channel (Fig. 7E). We also detected a novel π-π (stacking) interaction between Y37 of ImKTX58 and H404 of KV1.3 channel (Fig. 7F), which has not been reported in any known toxins blocking KV channels. Of note, besides H404, the D402, M403, P405, and V406 in D chain of KV1.3 channel are also in a close distance to Y37 of ImKTX58 within the range of 4Å (Fig. 7F).
Discussion
Toxic animals like scorpions and spiders could secrete venoms which contain numerous ion channel regulatory peptides to capture prey and defend themselves. 2621 species of scorpions have been identified and listed on the scorpion files (https://www.ntnu.no/ub/scorpion-files/) website, and about 50–100 different toxin peptides exist in each scorpion venom, which are important resources for developing selective and potent Na+ and K+ channel inhibitors (Liu et al., 2009; Han et al., 2011). But the numbers of scorpion species being used to explore bioactive peptides are scarce because only about 0.4–0.5% of toxin peptides have been reported and characterized thoroughly for now (Liu et al., 2009; Han et al., 2011). The toxin polypeptides from Hainan I. maculatus are rarely explored and far less than that of Buthus martensii Karsch. In this study, we identified the ImKTX58 gene from a cDNA Library of Hainan I. maculatus venom glands and used the prokaryotic expression system to express the polypeptides coded by this gene. We tested the bioactivity of therImKTX58 with whole-cell patch-clamp technique and found that this polypeptide has an inhibitory effect on KV1.3 channel endogenously expressed in Jurkat T cells and heterologously expressed in HEK293T cells. Further electrophysiological studies revealed that ImKTX58 peptide had a high blocking selectivity on KV1.3 over other K+ channels such as KV1.1, KV1.2, Kv1.5, SK2, SK3, and BK channels, validating ImKTX58 as a selective KV1.3 channel blocker.
Sequence alignment analysis revealed that the ImKTX58 polypeptide consisting of 38 amino acid residues belongs to the α-KTX subfamily of scorpion peptide toxin, and its typical cysteine-stabilized–α/β molecular skeleton structure is maintained by three disulfide bonds (Giangiacomo et al., 2004; Zhao et al., 2015). According to the previous studies on structure and function of α-KTX, their interaction surfaces with KV channels are mainly laid in three sites: β-strands, α/β-turn, and β-turn between the second and the third β-strands in C-terminus (Giangiacomo et al., 2004). Our primitive results of molecular modeling showed that the C terminal of ImKTX58 contains two reverse-parallel β-strands that are made up of 16 amino acids. Among these 16 amino acids, three cysteine are key to form disulfide bonds and will destroy the basic structure if mutated to alanine. Two glycine and one alanine were not mutated because they have the same property. We applied alanine scanning mutagenesis on the left 10 sites. K33A mutant could not be expressed. T35A and W38A mutants have totally different secondary structure with WT and did not show bioactivities because they could not inhibit KV1.3 channel even in 10 μM concentration (data not shown). The 7 mutants showed in the results keep the secondary structure with WT ImKTX58 and their electrophysiology data could be used to explain the results of computer simulation. Most toxin peptides use their β strands in C-terminus as the active surface to interact with KV1 channels and block the pore of KV1 channels through the functional dyad consisting of a basic amino acid residue (usually a lysine) and a strong hydrophobic aromatic amino acid residue (usually a tyrosine) like Lys27 and Tyr36 for Charybdotoxin (ChTX) or Lys22 and Tyr23 for Stichodactyla helianthus toxin (Giangiacomo et al., 2004; Zhao et al., 2015). ImKTX58 reported in this study also has Lys28 and Tyr37 residues that may compose the functional dyad and become the most important two amino acid residues to interact with KV1.3 channel (Liu et al., 2009; Han et al., 2011). Based on the X-ray structure of ChTX in complex with KV2.1 paddle–KV1.2 chimera channel, the lysine residue protrudes in the selectivity filter of the KV channel pore to block the K+ fluxes (Banerjee et al., 2013). Our electrophysiological results confirmed that Lys28 of ImKTX58 is the most important amino acid residue that mediate the interaction of ImKTX58 with Kv1.3 channel. The IC50 was decreased almost 1,000-fold when the Lys28 was mutated to alanine. Furthermore, the results of computer simulation revealed that the Lys28 of ImKTX58 interacts with the conserved GYG motif in the filter region of KV1.3 channel. These results are consistent with previous studies showing that the conserved lysine residue as a pore blocking site in β-strand of the toxin interacting with KV channels (Giangiacomo et al., 2004; Banerjee et al., 2013). Besides the lysine residue, another residue in this dyad—tyrosine—usually interacts with the outer vestibules of the KV channels to stabilize the combination between the toxin and the channel (Giangiacomo et al., 2004). Although our electrophysiological studies showed that the role of Tyr37 in mediating the interaction between ImKTX58 polypeptide and KV1.3 channel is weaker than expected, the novel π-π (stacking) interaction existed between Y37 benzene ring of ImKTX58 and H404 imidazole ring in A chain of KV1.3 channel might be an important structural basis for the high affinity binding of ImKTX58 to KV1.3 channel. Although the π-π interaction has not been reported in known α-KTX toxins, H404 residue of KV1.3 channel was reported to be critical for the interaction between multiple channel blockers such as Tetraethylammonium (TEA), ChTX, and ADWX-1 with KV1.3 outer vestibule regions (Naranjo and Miller, 1996; Bretschneider et al., 1999; Yin et al., 2008). Further experiments using mutant cycle analysis are needed to explore the interaction of Y37-H404 site, which might be a key site for increasing the binding potency of ImKTX58 with KV1.3 channel.
Based on our electrophysiological results, R24 and N31 of ImKTX58 play more important roles in modulating the KV1.3 channel compared with Y37 because mutating these two residues to alanine would decrease the IC50 on KV1.3 channel for 91.18- and 154.87-fold, while Y37A mutation only decrease the IC50 for 47.67-fold. These two residues could form ionic and hydrogen bonds with KV1.3 channel respectively. Interestingly, previous studies on α-KTX toxins also showed that the “Asn30” residue in the β-turn binds to the receptor residues in KV1.3 channels with hydrogen bonds and determines the affinity of multiple toxins to KV1.3 channel (Schroeder et al., 2002; Giangiacomo et al., 2004). We suspect that N31 laying in the β-turn of ImKTX58 may have similar function of “Asn30” residue in other KV1.3 toxins and determine the specificity of interaction between ImKTX58 and KV1.3 channel. Studies on ChTX and Agitxoxin-2 (AgTX-2) showed that their Arg residues (Arg24 for ChTX and Arg25 for AgTX2) in the α/β-turn of the toxins made electrostatic interaction with acidic residues like Asp (D) in KV1 channel pore and determine the selectivity of the toxins to KV1 channels (Goldstein et al., 1994; Hidalgo and MacKinnon, 1995). Our model showed the same interaction between Arg24 in α/β-turn and Asp402 in KV1.3 channel. Moreover, the S378, S379, and G380 in C chain of KV1.3 channel interact Arg24 with ionic or hydrogen bonds, which are different from that of the classic α-KTX scorpion toxin polypeptides (Giangiacomo et al., 2004; Yin et al., 2008). Therefore, our studies on ImKTX58 further enriches the diversity of the interactions between scorpion venom peptides and ion channels, which provides a new theoretical basis for the structural modification of scorpion venom peptides targeting the ion channels. Combined with X-ray and Cryogenic Electron Microscopy (Cryo-EM) study on protein structures, we expect to confirm the computer simulation of interactions between ImKTX58 and KV1.3 and design new ImKTX58 peptides to reach a higher blocking selectivity and efficiency (Chen et al., 2010; Banerjee et al., 2013).
The number of KV1.3 channels on the membrane of activated TEM cells is significantly increased in autoimmune diseases (Wulff et al., 2003; Zhao et al., 2015). Selective blocking KV1.3 channels can effectively inhibit the activation of TEM cells and the occurrence of autoimmune diseases (Matheu et al., 2008; Pérez-Verdaguer et al., 2016). Therefore, KV1.3 channel has been considered as an effective drug target for the treatment of autoimmune diseases and some drugs targeted KV1.3 are processed in clinical test (Al Musaimi et al., 2018). Most of these drugs are designed and developed based on peptide toxins derived from animals (Al Musaimi et al., 2018; Wulff et al., 2019). Since ImKTX58 peptide can selectively and efficiently block KV1.3 channel, it has the potential to be used to study the structure and function of Kv1.3 channels and also serve as a template for the development of KV1.3 blockers. However, compared with ADWX-1, O. scrobiculosus toxin-1, and other modified KV1.3 channel blockers like S. helianthus toxin-186, which inhibits KV1.3 channel in a picomolar concentration, the bioactivity of ImKTX58 peptide still has a large improvement space (Yin et al., 2008; Bhuyan and Seal, 2015; Zhao et al., 2015). We will perform the structural optimization based on the C-terminus amino acid residues of ImKTX58 to engineer new ImKTX58 peptides with significantly improved blocking efficiency and specificity as well as the half-life in circulation so that they can be used in animal disease models in future (Pennington et al., 2009; Edwards et al., 2014; Zhu et al., 2015).
Authorship Contributions
Participated in research design: Yin.
Conducted experiments: Zhang, Zhao, Yang, Lan, Y. Li, Xiao, Yu, Z. Li, Zhou, Wu, Cao, Yin.
Contributed new reagents or analytic tools: Zhang, Zhao, Yang, Lan, Wu, Cao, Yin.
Performed data analysis: Zhang, Zhao, Yang, Lan, Y. Li, Yin.
Wrote or contributed to the writing of the manuscript: Yin, Zhao.
Footnotes
- Received December 22, 2021.
- Accepted June 19, 2022.
This work was supported partly by National Natural Sciences Foundation of China [Grant 81373379] and [Grant 81641186] and National Key R and D Program of China [Grant 2019YFC1712402] (S.Y.), National Natural Sciences Foundation of China [Grant 32000685] and Natural Sciences Foundation of Hubei Province [Grant 2020CFB348] (Q.Z.), and Fundamental Research Funds for the Central Universities, South-Central University for Nationalities [CZZ19005] (S.Y.) and [CZQ20009] (Q.Z.).
All authors declare no conflicts of interest.
↵1 X.Z. and Q.Z. contributed equally to this work.
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- ADWX-1
- autoimmune drug from Wenxin group
- APC
- anterior piriform cortex
- BK
- large conductance voltage and Ca2+-activated potassium
- ChTX
- Charybdotoxin
- DRG
- dorsal root ganglion
- GSH
- glutathione
- GST
- glutathione S-transferase
- HPLC
- high performance liquid chromatography
- ImKTX58
- number 58 potassium channel toxin peptide identified from the venom gland cDNA library of Isometrus maculatus
- ImKTX88
- number 88 potassium channel toxin peptide identified from the venom gland cDNA library of Isometrus maculatus
- α-KTX
- alpha-potassium channel toxin peptides
- Kv
- voltage-gated potassium
- LmKTX10
- number 10 potassium channel toxin peptide identified from the venom gland cDNA of Lychas mucronatus
- MALDI-TOF-MS
- matrix assisted-laser-desorption/ionization time-of-flight mass spectrometry
- MD
- molecular dynamic
- SK2 channel
- small conductance calcium-activated potassium channel 2
- SK3 channel
- small conductance calcium-activated potassium channel 3
- TEM cell
- effector memory T cell
- WT
- wild-type
- Copyright © 2022 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.