Abstract
Aberrant dopamine (DA) signaling is associated with several psychiatric disorders, such as autism, bipolar disorder, addiction, and Parkinson’s disease, and several medications that target the DA transporter (DAT) can induce or treat these disorders. In addition, psychostimulants, such as cocaine and D-amphetamine (AMPH), rely on the competitive interactions with the transporter’s substrate binding site to produce their rewarding effects. Agents that exhibit noncompetitive, allosteric modulation of DAT remain an important topic of investigation due to their potential therapeutic applications. We previously identified a novel allosteric modulator of human DAT, KM822, that can decrease the affinity of cocaine for DAT and attenuate cocaine-elicited behaviors; however, whether DAT is the sole mediator of KM822 actions in vivo is unproven given the large number of potential off-target sites. Here, we provide in silico and in vitro evidence that the allosteric site engaged by KM822 is conserved between human DAT and Caenorhabditis elegans DAT-1. KM822 binds to a similar pocket in DAT-1 as previously identified in human DAT. In functional dopamine uptake assays, KM822 affects the interaction between AMPH and DAT-1 by reducing the affinity of AMPH for DAT-1. Finally, through a combination of genetic and pharmacological in vivo approaches we provide evidence that KM822 diminishes the behavioral actions of AMPH on swimming-induced paralysis through a direct allosteric modulation of DAT-1. More broadly, our findings demonstrate allosteric modulation of DAT as a behavior modifying strategy and suggests that Caenorhabditis elegans can be operationalized to identify and investigate the interactions of DAT allosteric modulators.
SIGNIFICANCE STATEMENT We previously demonstrated that the dopamine transporter (DAT) allosteric modulator KM822 decreases cocaine affinity for human DAT. Here, using in silico and in vivo genetic approaches, we extend this finding to interactions with amphetamine, demonstrating evolutionary conservation of the DAT allosteric site. In Caenorhabditis elegans, we report that KM822 suppresses amphetamine behavioral effects via specific interactions with DAT-1. Our findings reveal Caenorhabditis elegans as a new tool to study allosteric modulation of DAT and its behavioral consequences.
Introduction
Dopamine (DA) is a conserved neurotransmitter that regulates a variety of complex behaviors across phylogeny (McDonald et al., 2007; Yamamoto & Vernier, 2011). The DA transporter (DAT) regulates DA signaling by restricting DA actions spatially and temporally (Kaya et al., 2018; J. Zhu & Reith, 2008; S. Zhu et al., 2015). Altered DAT function is associated with multiple brain disorders including schizophrenia, Parkinsonism/dystonia, autism, and addiction (Aguilar et al., 2021; Belovich et al., 2021; Bowton et al., 2014; Del Campo et al., 2011; DiCarlo et al., 2019; Hamilton et al., 2013; Hornykiewicz, 2006; Keiflin & Janak, 2015; Kurian et al., 2009; Mazei-Robison et al., 2005; Salatino-Oliveira et al., 2018; J. Zhu & Reith, 2008).
Our understanding of DAT function has been facilitated by the elucidation of the structures of bacterial homologs (Yamashita et al., 2005), the Drosophila DAT (Penmatsa et al., 2015), and several mammalian members of the same transporter family (Coleman et al., 2016; Cuboni & Hausch, 2014). Computational modeling has also improved our understanding of the mechanism of function of DAT (Beuming et al., 2008; Cheng et al., 2015a; Cheng & Bahar, 2015; Kaya et al., 2018; Khelashvili et al., 2015). These studies agree on a central orthosteric site, S1, that binds the substrate DA and competitive inhibitors/substrates such as cocaine and amphetamine (APMH) (Aggarwal et al., 2021; Cheng & Bahar, 2019). Some studies have also suggested the existence of allosteric sites in the extracellular vestibule of DAT (Aggarwal & Mortensen, 2017; Navratna et al., 2018; Zhen & Reith, 2016) in the related serotonin transporter (Chen et al., 2005; Coleman et al., 2016; Niello et al., 2020; Plenge et al., 2020, 2021) and in the bacterial homolog LeuT (Beuming et al., 2008; Cheng & Bahar, 2013; Shan et al., 2011; Shi et al., 2008; Zhao et al., 2011). We have previously used structure/function studies to identify one allosteric site that we termed A2 (Aggarwal et al., 2019, 1). We identified a specific compound, KM822, that interacts with this site and have demonstrated that it interferes with the interaction of cocaine with DAT and attenuates cocaine-elicited locomotion in planaria (Aggarwal et al., 2019).
Drugs of abuse, such as cocaine and AMPH, act by targeting DAT, leading to substantial elevation of extra-synaptic DA levels (Belovich et al., 2021; Di Chiara & Imperato, 1988; Mayer et al., 2021; Mortensen & Amara, 2003; Saunders et al., 2000; J. Zhu & Reith, 2008). Recent reports suggest that mechanisms that govern human addiction are phylogenetically conserved (Engleman et al., 2016). The nematode Caenorhabditis (C.) elegans has a simple nervous system with highly conserved genes that regulate neuronal development, maintenance, and function (Hobert, 2010; McDonald et al., 2006; Refai et al., 2013; Serrano-Saiz et al., 2017; Sulston et al., 1975). We and others have made use of the nematode model to study DAT-dependent DA signaling (Bermingham et al., 2017; Carvelli et al., 2010; J. A. Hardaway et al., 2012; Nass et al., 2002; Refai & Blakely, 2019; Robinson et al., 2019).
Mutations in C. elegans DAT/DAT-1 result in a robust, conditional immobility phenotype that we designated swimming-induced paralysis (Swip), where animals lose their natural ability to swim in water (McDonald et al., 2007). Treatment of worms with the DAT-1 competitive substrate and DA releaser AMPH (Carvelli et al., 2010) or the high-affinity DAT-1 inhibitor nisoxetine (Bermingham et al., 2016), results in Swip that can be blocked by genetic or pharmacological blockade of DOP-3 (Carvelli et al., 2010; Refai & Blakely, 2019). Moreover, Swip phenotype has proven effective in studying the structural and functional characteristics of the human DAT (hDAT) (McDonald et al., 2007; Nass et al., 2005; Robinson et al., 2017).
Here, we capitalize on the power of a C. elegans model to characterize the action and specificity of the allosteric DAT modulator KM822 in vivo. Our data from molecular modeling studies support the binding of KM822 to a similar allosteric site in DAT-1 as in hDAT. Our pharmacological studies demonstrate that KM822 can both antagonize DAT-1 and reduce the inhibitory actions of AMPH in vitro. Consistent with these findings, KM822 attenuates the ability of AMPH to induce Swip at concentrations that do not impact DAT-1. Follow-up genetic and pharmacological studies support the targeting of DAT-1 by KM822 over other AMPH targets in vivo. We discuss our findings with respect to use of the worm model as a model for further evaluation of KM822 and other DAT allosteric modulators, which provide a novel therapeutic strategy for the treatment of disorders that feature DAT signaling perturbations.
Materials and Methods
Reagents and Drugs
Radiolabeled substrates [3H]-dopamine (32.6 Ci/mmol) and [3H]-serotonin (23.9 Ci/mmol) were purchased from PerkinElmer (Boston, MA, USA). Cell culture media and supplements, including penicillin/streptomycin, Dulbecco’s phosphate-buffered saline (DPBS), Dulbecco’s modified Eagle’s medium with glucose, and scintillation fluid, were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Transfection reagents TransIT-LT1 were from Mirus Bio, LLC (Madison, WI, USA). Reagents for uptake assays and non-radiolabeled substrates were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Structural Models of C. elegans DAT
The structural models of C. elegans DAT-1 (residues R40 to K594; UniProt ID Q03614)) in the outward-facing open (OFo) and inward-facing open (IFo) states were generated using the OFo Drosophila melanogaster DAT structure [Protein Data Bank (PDB): 4M48] (Penmatsa et al., 2013) and IFo human serotonin transporter (PDB: 6DZZ) (Coleman et al., 2019) as the template, respectively. We used the homology modeling protocol described earlier (Cheng & Bahar, 2015). Briefly, an ensemble of homology models was generated using MODELER (Fiser & Šali, 2003), and the model with the lowest (MODELER objective function) score was selected for further docking simulations.
Docking Simulations
The molecular structure of KM822 was taken from a previous study (Aggarwal et al., 2019). The binding sites and poses of KM822 onto C. elegans DAT (DAT-1) were assessed using the protein-ligand docking software AutoDock (Trott & Olson, 2009). Docking simulations were performed following the previous protocols (Cheng et al., 2015b). Briefly, a Lamarckian genetic algorithm with default parameters was employed, with the maximal number of energy evaluations set to 2.5 × 107. The binding energy was estimated from the weighted average of multiple binding poses at a given site observed in 100 independent runs.
Cell Culture and Transfections
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (3.5-g/L glucose) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C with 5% CO2. For transient transfections, COS-7 cells were transfected using the TransIT-LT1 transfection reagent (Mirus Bio, LLC, Madison, WI, USA).
Transport Inhibition Assays Using COS-7 Cells
Transiently transfected COS-7 cells (expressing DAT-1) were plated in 24-well plates. Uptake experiments were performed 2 days later. The medium was removed, and the cells were washed with PBS (137 mM of NaCl, 2.7 mM of KCl, 4.3 mM of Na2HPO4, and 1.4 mM of KH2PO4, pH 7.4) containing 0.1 mM of CaCl2, 1 mM of MgCl2, 5 mM of RO 41-0960, and 100 mM of ascorbic acid. Following washing, the cells were incubated for 10 minutes with various concentrations of KM822, and the uptake was initiated by adding [3H]DA to a final concentration of 50 nM. Uptake was allowed to continue for 10 minutes at room temperature and was terminated by washing twice with ascorbic. Cells were solubilized in a scintillation cocktail and counted on a microplate scintillation and luminescence counter (PerkinElmer, Waltham, MA, USA). A Hill equation was fitted to data by a nonlinear regression analysis to obtain the IC50 values.
C. elegans Strains
C. elegans strains were cultured on OP50 or NA22 bacterial lawns and maintained at 13–20°C using standard protocols (Brenner, 1974). Worm strain N2 Bristol was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) and used as the wild-type strain. The dat-1(ok157) strain was used as the DAT-1 loss of function allele, whereas a loss of function allele of the LGC-55 ion channel (lgc-55(n4331)) and the double KO strain dat-1; lgc-55 (a kind gift from Dr. Lucia Carvelli, Florida Atlantic University) were used to assess a role of LGC-55 versus DAT-1 dependence of KM822 inhibition of AMPH actions.
C. elegans Swip Assays
Swip assays were performed as previously described (J. A. Hardaway et al., 2012; McDonald et al., 2007). Briefly, worms were grown on NA22 plates, with synchronization achieved by lysis of gravid adults using 4%(ml) hypochlorite treatment. Synchronized L1 animals were plated into OP50 plates until the L4 stage. All Swip assays were initiated by placing ten L4 hermaphrodites into a well of 100 μl of distilled water (note Swip does not occur in isotonic M9 medium) containing vehicle, plus or minus drug. Animals were scored as the number of paralyzed versus swimming worms after a 10-minute incubation. For each genotype and/or treatment, eight wells were scored, and every experiment was repeated on at least three separate days for an n = 24. For drug treatments, KM822 was dissolved in DMSO to generate a 50 mM stock solution, which was used to generate dilutions in distilled water for Swip assays. The final concentration of DMSO in drug solutions of 50 μM, 250 μM, and 500 μM of KM822 equals 0.1%, 0.25%, and 0.5%, respectively. Vehicle controls used the same DMSO concentrations in the absence of drug. AMPH (Sigma-Aldrich, St. Louis, MO) was prepared as 50 mM–100 mM stock solutions and used at serial dilutions at 300 μM. β-phenylethylamine (Sigma-Aldrich, St. Louis, MO; a gift from Lucia Carvelli, Florida Atlantic University) was prepared as a stock solution in distilled water at 10 mM and used at a final concentration of 1 mM, as previously described (Safratowich et al., 2014). For the drug mixtures, AMPH was added to KM822 or β-PEA final concentration solutions, mixed vigorously by vortexing and shaking, and used immediately after preparation. Drug solutions were used as a swimming medium for the total time of the Swip assay, to achieve acute treatment.
Statistical Analyses
Swip data were analyzed statistically and graphed using Prism version 7.0 (GraphPad, Inc., La Jolla, CA). For the Swip assays, the sample size for each group is 10 animals per well, for eight wells tested per assay (n = 80) per genotype, following the previously described protocol (see above). Unless otherwise stated, data were analyzed by Student’s t tests, one-way ANOVAs and two-way ANOVAs followed by Sudak, Dunnet’s post-hoc, or Bonferroni multiple comparison tests. Statistical significance was considered achieved for P < 0.05 in all cases. Error bars were plotted to represent the mean with SD, unless otherwise mentioned. Statistical inter-group comparisons were decided after data were observed. Our experiments were carried out in an exploratory manner; therefore, the calculated P values shall be interpreted as such, rather than a hypothesis-testing approach.
Results
Computational modeling and functional uptake assays indicate KM822 interacts with DAT-1
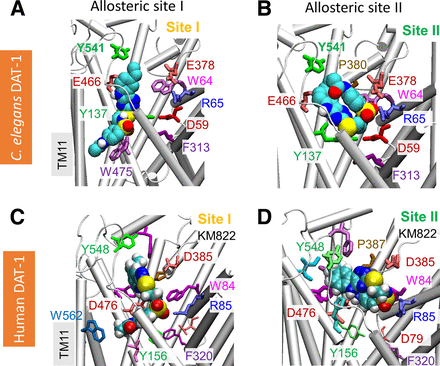
Structural models of DAT-1 in the OFo and IFo states were generated using the OFo Drosophila melanogaster DAT (PDB: 4M48) (Penmatsa et al., 2013) and IFo human serotonin transporter (PDB: 6DZZ) (Coleman et al., 2019) as the template, respectively. DAT-1 has 47% sequence identity to hDAT and human serotonin transporter. In DAT-1, the conserved EC gating residues include R65-E466 and Y137-F313, whose homologous EC gates in hDAT are R85-D476 and Y156-F320 (see Fig. 1). The potential binding poses of KM822 and its interactions with DAT-1 in different conformation states were analyzed by a series of docking simulations. Similar to that observed in hDAT (Aggarwal et al., 2019, 2021), the most favorable binding site for KM822 bound to DAT-1 was within the EC vestibule in the OFo conformer, and no high affinity binding site was observed in the case of the IFo conformer. Therefore, we propose that KM822 would predominantly bind to the EC vestibule of OFo DAT-1. Two slightly different binding poses of KM822 (I and II) were identified within the A2 site of DAT-1 (Fig. 1), in agreement with those observed in MD simulations of KM822 binding to hDAT (Aggarwal et al., 2019, 2021). In DAT-1, KM822 pose I site is composed by e.g., Y137, E466, W475, and Y541 (Fig. 1a), which is comparatively deeper inside the EC vestibule than the pose II site. The KM822 pose II site is near the EC vestibule entrance (Fig. 1A), composed of e.g., W64, R65, E378, P380, E466, and Y541 (Fig. 1B). Overall, docking simulations indicated that KM822 exhibited high-affinity binding (−8.1±1.1 kcal/mol) to the A2 pocket located in close proximity to salt bridge formed by amino acids R65 and E466 (Fig. 1). The homologous salt bridge forms an external gate/lid in the occluded conformation transiently stabilized during the transport cycle of hDAT (review see, (Cheng & Bahar, 2019)), suggesting that KM822 through its interaction with the A2 allosteric site is centrally positioned to interfere with conformational changes during the transport cycle, altering the interactions of ligands with the orthosteric site.
KM822 binds to Caenorhabditis elegans dopamine transporter-1 at an allosteric site that closely overlaps with the KM822-binding site onto human dopamine transporter. Structural modeling and docking analysis suggest KM822 (space-filling; cyan, with N-, O-, and S-atoms in blue, red, and yellow, respectively) may bind to dopamine transporter-1 with two different poses (panels A and B), comparable to those at the allosteric site A2 previously described for hDAT (panels C and D), see (Aggarwal et al., 2021). Residues that coordinate KM822 in all four cases are shown in sticks and labeled.
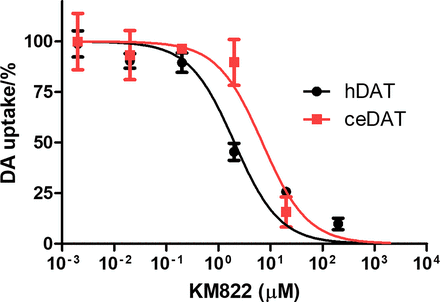
Functional studies on DA uptake confirmed the computational predictions as concentration-response inhibition curves showed that KM822 inhibited the DAT-1 consistent with a single binding site, albeit with a higher IC50 compared with KM822 inhibition of hDAT. The IC50 for inhibiting DAT-1 was 10.4 μM; 95% CI [6.3; 17.0] versus 2.2 μM; 95% CI [1.6; 3.2] for hDAT (Fig. 2). These findings encouraged us to determine whether KM822 affects DAT-1 function or its interactions with an orthosteric inhibitor in vivo.
KM822 suppresses the activity of both human and Caenorhabditis elegans dopamine transporters (DATs). Radioactive dopamine transport inhibition assay using KM822 against human (hDAT) and Caenorhabditis elegans (DAT-1) DAT in transiently transfected COS-7 cells. KM822 IC50 mean values for hDAT and DAT-1 are 2.2 μM, 95% confidence interval [1.6; 3.2] and 10.4 μM, 95% confidence interval [6.3; 17.0], respectively. Nonlinear regression was used to obtain a curve consistent with a single binding site. IC50 mean values and corresponding 95% confidence intervals are calculated using an average of five independent experiments.
KM822 Interferes with the Interaction of DAT-1 with AMPH
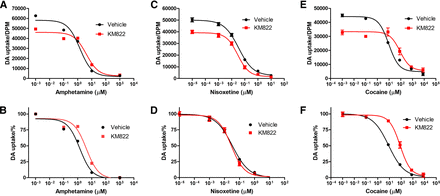
In our previous studies, we found that KM822 interferes with the interaction of cocaine with hDAT, reducing the affinity of cocaine (Aggarwal et al., 2019). Here, we studied the effect of KM822 on the interaction of the two psychostimulants, AMPH and cocaine, with DAT-1. Additionally, we tested nisoxetine as, unlike cocaine, it exhibits both a highly potent and DAT-specific ability to induce Swip (Bermingham et al., 2016; Carvelli et al., 2010; Valladares et al., 2021). First, we performed in vitro concentration-response DA transport inhibition assays in transfected COS-7 cells using AMPH, cocaine, and nisoxetine in the absence or presence of 20 µM of KM822 (Fig. 3). Our results demonstrate that KM822 shifts the concentration-response curve of AMPH-mediated inhibition to the right, indicating that KM822 affects AMPH interactions with DAT-1 (Fig. 3A, B). In contrast, little or no KM822 effect was observed on nisoxetine (Fig. 3C, D). The statistically significant effect of KM822 on the interaction of AMPH with DAT-1 is similar to what we observed previously with the interaction of hDAT with cocaine (Aggarwal et al., 2019). Similar to what we found in those previous studies, we observed that KM822 also shifts the concentration-response curve of cocaine-mediated inhibition of DAT-1 to the right (Fig. 3E, F). Taken together, these results support the hypothesis that KM822 interacts with the A2 allosteric site of DAT-1 in a manner that can attenuate the action of psychostimulants like cocaine and AMPH, reinforcing our decision to examine potential in vivo consequences of this.
KM822 affects dopamine transporter-1 interactions with amphetamine (AMPH) but has no effect on nisoxetine or cocaine actions. Two depictions of each representative dopamine uptake experiment are displayed. One of actual dopamine uptake (A, C, and E) and one with uptake values normalized relative to vehicle control (B, D, and F). (A, B) Radioactive dopamine (DA) transport inhibition assay of AMPH in the absence and presence of 20 µM of KM822 in dopamine transporter-1-transfected COS-7 cells. IC50 mean values of AMPH are 1.1 μM, 95% confidence interval (CI) [0.84; 1.4] for the vehicle and 3.4 μM, 95% CI [2.9; 3.9] in the presence of 20 µM of KM822 (**, p < 0.01). The concentration-response of AMPH-mediated inhibition was shifted to the right due to KM822 treatment. (C, D) Radioactive DA transport inhibition assay of nisoxetine in the absence and presence of 20 µM of KM822 in dopamine transporter-1-transfected COS-7 cells. IC50 mean values of nisoxetine in the absence and presence of 20 µM of KM822 are 0.04 μM, 95% CI [0.029; 0.055] and 0.03 μM, 95% CI [0.021; 0.042], respectively. No statistically significant change was observed in nisoxetine-mediated inhibition. (E, F) Radioactive DA transport inhibition assay of cocaine in the absence and presence of 20 µM of KM822 in dopamine transporter-1-transfected COS-7 cells. IC50 mean values of cocaine are 9.6 μM, 95% CI [8.1; 11] for the vehicle and 128 μM, 95% CI [61; 268] in the presence of 20 µM of KM822 (*, p < 0.05). The concentration-response of cocaine-mediated inhibition was shifted to the right due to KM822 treatment. IC50 mean values and 95% confidence intervals were calculated using the average of three independent experiments. Statistical analysis was performed using Student’s t test.
Acute KM822 Treatment Suppresses AMPH-induced Swip
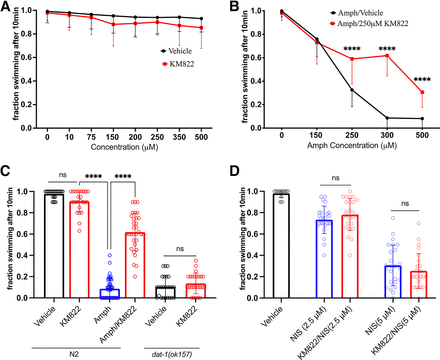
The effects of KM822 on AMPH-binding to DAT-1 in vitro suggested that KM822 may limit the action of AMPH in producing Swip, analogous to our findings that KM822 can diminish the actions of AMPH in the planeria model. As discussed earlier, the swimming behavior of C. elegans is tightly controlled by the activity of DAT-1, where loss-of-function mutations in DAT-1 result in the Swip phenotype (McDonald et al., 2007). As with a loss-of-function dat-1 allele, wild-type animals incubated in water with specific concentrations of AMPH exhibit Swip within a few minutes of incubation (Carvelli et al., 2010; Valladares et al., 2021). First, we analyzed the swimming activity of wild-type N2 animals in solutions containing increasing concentrations of KM822, ranging between 10 and 500 μM. Swip data recorded after 10 minutes of incubation in KM822 solutions indicated no statistically significant effect on swimming behavior of N2 (Fig. 4A). In contrast, and as expected, worms treated with AMPH demonstrated concentration-dependent induction of Swip up to 300µM, where a ceiling effect was observed (Fig. 4B). In contrast, animals treated with an AMPH/KM822 mixture exhibited clearly higher levels of swimming than observed with AMPH alone (Fig. 4B). The effect of KM822 on AMPH-induced Swip reached its peak at 300 μM AMPH and diminished at higher concentrations (500 μM AMPH), possibly due to competition between the two drugs for the target (Fig. 4C). Therefore, we used the concentrations 300 μM AMPH and 250 μM KM822 for subsequent experiments.
KM288 suppresses amphetamine (AMPH)-induced swimming-induced paralysis (Swip) in wild-type Caenorhabditis elegans: (A) concentration-response profile of wild-type (N2) to KM822 treatments indicating no statistically significant change in the animals’ swimming behavior (N ≥ 240). No statistical significance was observed comparing the swimming behavior of KM822 to vehicle treated animals of the same concentration point (P value < 0.05; F-value = 0.4368). Data were analyzed by a two-way ANOVA with Bonferroni multiple comparison tests between KM822 and vehicle treatments at each concentration. (B) KM288 (250 µM) suppresses AMPH-induced Swip phenotype at various AMPH concentrations. Swip assays were performed for animals incubated in distilled water solutions containing the depicted concentrations of AMPH or AMPH + KM822 (250 µM), N ≥ 240. Data were analyzed by a two-way ANOVA with Bonferroni multiple comparison tests, comparing AMPH and AMPH + KM822 treatments at each concentration point. Statistically significant differences were observed at 250 µM, 300 µM and 500 µM AMPH concentrations (**** = P value < 0.0001; F-value = 43.05). No statistically significant changes in the swimming profile of control or vehicle groups, as indicated in A (not shown for simplicity). (C) KM822 reversal of Swip is dopamine transporter-1 dependent. A representation of the 10-min Swip assay, depicting the swimming behavior of animals after 10 min of incubation in control (vehicle) or drug conditions (N ≥ 240). No statistically significant differences were observed comparing N2 in vehicle versus 250 μM KM822 conditions (ns, P value < 0.05). Whereas N2 animals incubated in 300 μM 0f AMPH demonstrated significant Swip paralysis compared to the 250 μM of KM822 and AMPH/KM822 mixture (**** = P value < 0.0001, F-value = 192.4). A dose of 250 μM of KM822 demonstrated no statistically significant effect on the Swip paralysis of dat-1 mutants compared to the condition (ns, P value < 0.05). Data were analyzed by a one-way ANOVA with Bonferroni multiple comparison tests. (D) KM822 has no statistically significant effect on nisoxetine-induced Swip at 2.5 μM and 5 μM of nisoxetine concentration (ns, P value < 0.05). Data were evaluated using one-way ANOVA or two-way ANOVA with selected Bonferroni’s multiple comparisons tests, comparing different treatments with (**** = P value < 0.0001; F-value = 156.4). N ≥ 240. For all data, means and SDs are displayed.
If the ability of KM822 to diminish AMPH-induced Swip is due to DAT-1 interactions, no reduction in Swip should occur in animals that lack DAT-1, i.e., in dat-1(ok157) mutants. Indeed, unlike KM822 effects on AMPH-induced Swip, we observed no ability of KM822 to diminish Swip in dat-1(ok157) animals (Fig. 4C). Additionally, and consistent with our in vitro pharmacological assays, KM822 displayed no statistically significant effect on nisoxetine-induced Swip, at either 2.5 μM or 5 μM concentrations (Fig. 4D).
KM822 Suppresses DAT-specific Actions of AMPH to Produce Swip
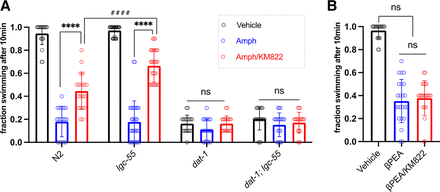
Although a substantial portion of AMPH-induced Swip has been found to be dependent of DAT-1 interactions, with effects attenuated when synthesis of DA is precluded or when expression of the DOP-3 receptor is prevented, a portion of AMPH-induced Swip has been reported to arise from the drug’s interaction with an amine-gated chloride channel LGC-55 (Safratowich et al., 2013, 2014). We therefore asked whether the loss of function strain lgc-55(n4331) alters the ability of KM822 to attenuate AMPH-induced Swip. We found that lgc-55 mutant animals display swimming behavior like that of N2 (Fig. 5A). Moreover, lgc-55 animals displayed an equivalent ability to support AMPH-induced Swip as N2. Importantly, KM822 suppressed AMPH-induced Swip in lgc-55 mutants even better than in N2. These effects indicate that the LGC-55 channel is not required for KM822 to diminish AMPH-induced Swip, in contrast to an essential requirement observed for DAT-1 (Fig. 4b, 5a). Indeed, loss of LGC-55 appears to render the effect of KM822 more penetrant (Fig. 5a), although the mechanism for this effect is unclear at present. Importantly, no ability of KM822 to rescue AMPH-induced Swip was evident in dat-1; lgc-55 mutants, consistent with DAT-1 but not LGC-55 as responsible for the AMPH-related actions of KM822. Consistent with this idea, the paralytic action of β-phenylethylamine, which mimics the effect of AMPH on C. elegans by activating the LGC-55 channels and induces DA signaling-independent Swip (Refai & Blakely, 2019), was insensitive to KM822 (Fig. 5B).
KM822 suppresses dopamine transporter (DAT)-specific swimming-induced paralysis (Swip) behavior, but not LGC-55 induced Swip. A) KM822 suppresses DAT-1 dependent Swip, but not LGC-55 induced Swip, in response to amphetamine (AMPH) treatment. Depicted are 10-min Swip assays for N2, lgc-55, dat-1, and dat-1; lgc-55 animals (N ≥ 240). KM822 suppresses AMPH-induced Swip in N2 animals and lgc-55 mutants (**** P value < 0.0001; F-value = 80.82), with KM822 suppression of AMPH-induced Swip found to be significantly greater in lgc-55 than in N2 background (#### P value < 0.0001; F-value = 80.82). KM822 has no statistically significant effect on AMPH-induced Swip of dat-1 and dat-1; lgc-55 mutant backgrounds (ns, P value < 0.05). Data were analyzed using two-way ANOVA with selected Bonferroni’s multiple comparisons tests, comparing KM822 treatments over conditions and genotypes. B) KM822 demonstrates no effect on β-phenylethylamine-induced Swip of N2 animals (ns, P value < 0.05). Swip data were analyzed using ordinary one-way ANOVA, with selected Bonferroni’s multiple comparisons tests, comparing different conditions with (**** = P value < 0.0001; F-value = 151.1). N ≥ 140. For all, data means and SDs are displayed.
Discussion
The results of our present study reveals an evolutionary conservation in the ability of KM822 to allosterically modulate DAT proteins, ranging from invertebrates to hDAT (Aggarwal et al., 2019). Our conclusion is supported by in silico structural modeling and docking analysis, in vitro pharmacological studies, and in vivo genetic and behavioral assays. Computational modeling of KM822 within DAT-1 shows that KM822 binds to the OFo of DAT-1 and preferentially occupies a region in the EC vestibule corresponding to the A2 pocket of hDAT. Both charged (e.g., E378 in DAT-1 and D385 in hDAT) and aromatic residues (e.g., W64 in DAT-1 and W85 in hDAT) within the allosteric binding site are conserved between hDAT and DAT-1. Consistent with what was found for hDAT, KM822 sits in the allosteric domain of DAT-1 close to the R65-E466 salt bridge and may thereby influence affinity of ligands bound at the orthosteric site by interfering with conformations needed to inhibit the transport cycle. Based on the computational predictions, future structure/functions studies will be important to identify specific residues within DAT-1 that are important for mediating the interaction with KM822. Determination of in vitro activity of KM822 in DAT-1-transfected COS-7 cells revealed that KM822 potently inhibits DAT-1 transport function as was previously observed for hDAT. Interestingly, KM822 effectively reduced the transport inhibition potency of AMPH- and cocaine-mediated DAT-1 function but had no effect on the potency of the DAT-1 inhibitor nisoxetine. These results are similar to our observation in previous studies, where we have shown that KM822 attenuated the inhibitory potency of cocaine in hDAT-mediated DA transport assay (Aggarwal et al., 2019). Collectively, these results prove that KM822 binds to an allosteric site of DAT-1 that is separate from the orthosteric binding site and interferes with the transport mechanism of DAT-1, most likely by inducing conformational changes in the transporter. In vitro studies show that KM822 has no effect on the inhibition potency of another orthosteric ligand, nisoxetine, in DAT-1 transfected cells, indicating that the attenuation of AMPH and cocaine activity in DAT-1 transfected cells by KM822 might not be due to a direct interference with ligand binding at the orthosteric site versus either interference at a more distal site or perturbations of conformational changes that must be achieved by the orthosteric ligand to achieve high affinity binding. Interestingly, the affinities for the two orthosteric inhibitors, cocaine and nisoxetine, are different with the higher affinity ligand nisoxetine not being affected by KM822 engagement of the allosteric site. Taken together, our findings demonstrate an evolutionary conservation of the allosteric pocket that likely provides the structures necessary for mediating allosteric effects, but also points to ligand-specific effects as KM822 affects the interaction of the orthosteric ligand cocaine but not nisoxetine with DAT. This furthermore opens the possibility of developing allosteric ligands with unique specificity and selectivity and with various transporter-modulating activities.
To determine whether findings with KM822 heterologous cell transfection of DAT-1 cDNA are relevant in vivo, we evaluated the ability of KM822 to diminish AMPH-induced Swip behavior in which nematodes deficient in DAT-1 expression are exposed to water. The Swip phenotype has been used extensively to investigate the structural, functional, and behavioral properties of DAT-1 in vivo and as a tool to support forward genetic studies to identify novel determinants of DA signaling and health (Bermingham et al., 2017; Carvelli et al., 2010; Gibson et al., 2018; J. Hardaway et al., 2015; J. A. Hardaway et al., 2012; Refai & Blakely, 2019; Robinson et al., 2019; Safratowich et al., 2013). The free-living C. elegans hermaphrodite possesses eight mechanosensitive DA neurons that inhibit motor neurons involved in determining the speed of crawling on solid surfaces or thrashing in liquid. Excess DA secretion, or reduced DA clearance via DAT-1 leads, when sustained, to Swip. Swip is triggered by excess DA release that can arise from mutations that induce excess DA neuron excitability (J. Hardaway et al., 2015) when DAT-1 is antagonized pharmacologically (e.g., with nisoxetine) (Bermingham et al., 2016) or when DAT-1 is genetically eliminated (McDonald et al., 2007). As a competitive substrate of DAT-1 with the ability to induce DAT-1-mediated DA efflux, AMPH also produces Swip (Carvelli et al., 2010; Safratowich et al., 2013, 2014; Valladares et al., 2021) (ref) via both DAT-1 dependent and independent mechanisms (McDonald et al., 2007; Safratowich et al., 2013).
Since tests of KM822 interaction with DA uptake assays in DAT-1-transfected cells revealed inhibitory activity, we expected the drug might induce Swip like the DAT-1 inhibitor nisoxetine. However, KM822 at the concentrations used had no effect on the swimming behavior of wild-type animals, suggesting that the drug’s allosteric interactions with DAT-1 are insufficient to produce Swip, or that the drug does not access the transporter in vivo, perhaps due to poor penetration across the worm cuticle. However, we found KM822 to be bioactive in vivo when tested in the context of AMPH-induced Swip. We have reported analogous observations previously in planarians, where KM822 did not impact locomotion in planarians, but effectively antagonized cocaine-induced stimulation of locomotion (Aggarwal et al., 2019). The observation that KM822 does not affect nisoxetine-induced Swip supports the in vitro results that showed that KM822 had no effect on DAT-1 interactions with nisoxetine. This further corroborates that the KM822 interaction with DAT-1 occurs either through an intermediate or because of binding to sites distinct from those of an orthosteric inhibitor. Since we see a shift induced by KM822 of AMPH potency in transfected cells not expected to harbor C. elegans DAT-1-interacting proteins, and we detect a conserved binding site for KM822 shared by nematodes and humans, we believe our findings are most consistent with a model whereby KM822 exhibits non-competitive inhibition of DAT-1 via allosteric modulation of DAT-1 function. Such observations have been widely reported in the past with several other DAT ligands with non-competitive mechanism of inhibition (Li et al., 2011; Loland et al., 2008; Schmitt & Reith, 2012) though to our knowledge, this is the first example of an interaction that results in modulated AMPH action.
Our findings indicate that KM822 specifically targets DAT-1 in vivo based on the fact that: (1) KM822 has no effect on Swip behavior induced by dat-1 KO mutants, and (2) the effect of KM822 on the AMPH-induced Swip is lost in dat-1 animals. However, since AMPH can induce Swip in a DAT-1-independent manner via activation of the amine-gated chloride channel LGC-55 (Safratowich et al., 2013, 2014), we needed to confirm that the action of KM822 to attenuate AMPH-induced Swip was not due to an interaction of KM822 on LGC-55. First, we showed that KM822 suppression of AMPH-induced Swip is not lost (rather increased) in animals that lack LGC-55. Second, we found that KM822 was ineffective in reducing Swip behavior triggered by β-phenylethylamine, a specific activator of LGC-55 that induces Swip independently of DA signaling (Refai & Blakely, 2019). Together, our data indicate that KM822 specifically targets DAT-1 and allosterically reduces AMPH interactions with the transporter as was found in transfected cells. Consequently, we suggest that KM822 action on DAT-1 is via the occupancy of the A2 allosteric site first identified in hDAT (Aggarwal et al., 2019, 2021). Identification of A2 site mutations that disrupt KM822 interactions with AMPH, but that fail to alter DAT-1 activity, and the introduction of these changes in vivo through CRISPR/Cas9 approaches can add further support for our conclusion. To conclude, by using both pharmacological and genetic approaches, our studies demonstrate that the behavioral effects of KM822 are mediated by a specific modulation of DAT-1. This finding adds to our previous studies in planarians, here adding strong evidence that in vivo allosteric inhibition of DAT can be realized. Furthermore, our findings support the use of the worm model, with its extensive battery of genetic tools, as a platform for the rapid screening of DAT allosteric inhibitors as well as in understanding their mechanisms of action. Further pursuit of these efforts may provide lead structures for the development of novel therapeutic agents.
Acknowledgments
The authors would like to acknowledge the outstanding Caenorhabditis elegans laboratory support provided by Sean Mellish in the Blakely laboratory. The authors also thank Dr. Lucia Carvelli at FAU for providing materials and strains used in this study.
Authorship Contributions
Participated in research design: Refai, Aggarwal, Cheng, Bahar, Blakely, Mortensen.
Conducted experiments: Refai, Aggarwal, Cheng, Gichi.
Contributed new reagents or analytic tools: Refai, Aggarwal, Cheng, Salvino, Bahar, Blakely, Mortensen.
Performed data analysis: Refai, Aggarwal, Cheng, Bahar, Mortensen.
Wrote or contributed to the writing of the manuscript: Refai, Aggarwal, Cheng, Bahar, Blakely, Mortensen.
Footnotes
- Received August 24, 2021.
- Accepted December 3, 2021.
This work was supported by the National Institutes of Health [Grants NIH-MH106912 and NIH-MH121453 to O.V.M. and P41 GM103712 to I.B.] and the Community Foundation of Palm Beach County to R.D.B.
The authors declare the following competing financial interest(s): KM822 and its analogs are listed in the US Patent 9616065, with O.V.M. and J.M.S. as named inventors. All other authors have no actual or perceived conflicts of interest with the contents of this article.
↵1 O.R. and S.A. contributed equally to this work.
Abbreviations
- AMPH
- amphetamine
- CI
- confidence interval
- DA
- dopamine
- DAT
- dopamine transporter
- IFo
- inward-facing open
- OFo
- outward-facing open
- Swip
- swimming-induced paralysis
- Copyright © 2022 by The American Society for Pharmacology and Experimental Therapeutics