Abstract
Phenobarbital (PB), a broadly used antiseizure drug, was the first to be characterized as an inducer of cytochrome P450 by activation of the constitutive androstane receptor (CAR). Although PB is recognized as a conserved CAR activator among species via a well-documented indirect activation mechanism, conflicting results have been reported regarding PB regulation of the pregnane X receptor (PXR), a sister receptor of CAR, and the underlying mechanisms remain elusive. Here, we show that in a human CAR (hCAR)-knockout (KO) HepaRG cell line, PB significantly induces the expression of CYP2B6 and CYP3A4, two shared target genes of hCAR and human PXR (hPXR). In human primary hepatocytes and hCAR-KO HepaRG cells, PB-induced expression of CYP3A4 was markedly repressed by genetic knockdown or pharmacological inhibition of hPXR. Mechanistically, PB concentration dependently activates hPXR but not its mouse counterpart in cell-based luciferase assays. Mammalian two-hybrid assays demonstrated that PB selectively increases the functional interaction between the steroid receptor coactivator-1 and hPXR but not mouse PXR. Moreover, surface plasmon resonance binding affinity assay showed that PB directly binds to the ligand binding domain of hPXR (KD = 1.42 × 10−05). Structure-activity analysis further revealed that the amino acid tryptophan-299 within the ligand binding pocket of hPXR plays a key role in the agonistic binding of PB and mutation of tryptophan-299 disrupts PB activation of hPXR. Collectively, these data reveal that PB, a selective mouse CAR activator, activates both hCAR and hPXR, and provide novel mechanistic insights for PB-mediated activation of hPXR.
Introduction
Phenobarbital (PB) is a commonly used medication for epilepsy management, which is known to accelerate the metabolism and clearance of many drugs and endogenous substances by upregulating the expression of numerous hepatic genes encoding drug-metabolizing enzymes and transporters, with CYP2B and CYP3A genes as its prototypical targets (Miles et al., 1988; Kakizaki et al., 2003). The highly pleiotropic responses to PB also include increased proliferation of endoplasmic reticulum, alteration of cell-cycle checkpoint controls, inhibition of apoptosis, and promotion of liver tumor development (Kitagawa et al., 1979; Feldman et al., 1980; Luisier et al., 2014). Since first reported as a metabolism inducer half a century ago (Remmer and Merker, 1963), PB has been used extensively as a research tool for biochemical and pharmacological investigations of liver drug metabolism and elimination. This inductive feature has also expanded the therapeutic scope of PB to the treatment of neonatal jaundice and other hyperbilirubinemia, in which PB increases hepatic bilirubin metabolism by inducing the expression of UDP-glucuronosyltransferase 1A1 (Ritter et al., 1999). The molecular mechanisms underlying PB-mediated gene transactivation started to rapidly unveil when the constitutive androstane receptor [(CAR), NR1I3] was functionally linked to PB-induced expression of CYP2B genes two decades ago (Honkakoski et al., 1998). In response to PB treatment, CAR is dephosphorylated at threonine-38 (Mutoh et al., 2009) by protein phosphatase 2A, which leads to translocation of CAR into the nucleus of hepatocytes. Subsequently, CAR forms a heterodimer with the retinoid X receptor and binds to PB response cis-acting elements located upstream of PB-inducible genes, leading to increased drug metabolism and clearance (Kawamoto et al., 1999; Negishi, 2017). The essential role of CAR in PB-mediated induction of CYP2B was firmly established by experiments using CAR-null mice, in which loss of CAR completely abolished PB induction of Cyp2b10 in the liver (Wei et al., 2000). Moreover, as a known nongenotoxic hepatocarcinogen, PB increases the incidence of liver tumors in mice by a mode of action involving CAR activation, and such tumor promotion effect was also eliminated in CAR-null mice (Yamamoto et al., 2004). Together, these findings suggest that the diverse actions of PB in mouse predominantly rely on the activation of CAR.
The pregnane X receptor [(PXR), NR1I2] is the closest relative of CAR on the nuclear receptor superfamily tree. As xenobiotic sensors, CAR and PXR coordinate a pleiotropic defensive mechanism by which activation of these receptors upregulates a spectrum of distinct and overlapping target genes (Xie et al., 2000; Faucette et al., 2007; Roth et al., 2008). Specifically, CAR and PXR can recognize and bind to response elements located upstream of each other’s target genes with different affinities (Faucette et al., 2006). CAR and PXR also display significant promiscuity in ligand recognition and share many common pharmacological modulators. For instance, the antimalarial artemisinin and the antipsychotic chlorpromazine are activators of both CAR and PXR (Burk et al., 2005; Faucette et al., 2007), while clotrimazole and 1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide exhibit potent activation of PXR but deactivation of CAR (Moore et al., 2000; Li et al., 2008). Significant species differences of CAR and PXR have also been documented, where some chemicals such as 1,4-bis[2-(3,5-dichlorpyridyloxy)]benzene and pregnenolone 16α-carbonitrile (PCN) are selective mouse CAR and mouse PXR (mPXR) activators (Kliewer et al., 1998; Tzameli et al., 2000), while others like 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO) and rifampicin (RIF) activate human CAR (hCAR) and human PXR (hPXR), respectively (Bertilsson et al., 1998; Maglich et al., 2003). In the case of PB, it has long been used as a model compound to investigate CAR-mediated gene transcription from rodents to humans. Complete loss of CYP2B/3A induction in CAR-null mice demonstrates that PB is a mouse CAR but not mPXR activator. However, the role of PB in hPXR activation has yet to be convincingly established and the underlying molecular mechanism remains unclear.
In the current study, we provide experimental evidence to show that PB activates hPXR through direct ligand binding. In hCAR-knockout (KO) HepaRG cells, PB robustly induces the expression of both CYP2B6 and CYP3A4, while further inhibition of hPXR fully abolished this induction. In mammalian two-hybrid assays, PB enhanced recruitment of steroid receptor coactivator-1 (SRC-1) to hPXR but not mPXR. In contrast to its indirect activation of CAR, surface plasmon resonance (SPR) binding affinity assays reveal that PB directly binds to hPXR. Using combined biologic and computational approaches, we also identify that tryptophan-299 (W299) in the ligand binding pocket of hPXR plays a key role in coordinating PB activation of hPXR.
Materials and Methods
Chemicals and Biologic Reagents.
PB, RIF, and CITCO were purchased from Sigma-Aldrich (St. Louis, MO). 4-((4-(Tert-butyl)phenyl)sulfonyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole (SPA70) was obtained from AK Scientific, Inc. (Union City, CA). Primers for real-time polymerase chain reaction (RT-PCR) were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The Dual-Luciferase Reporter Assay System was purchased from Promega (Madison, WI). Matrigel was obtained from BD Biosciences (Bedford, MA). Other cell culture reagents were purchased from Life Technologies (Grand Island, NY) or Sigma-Aldrich.
Plasmid Constructions.
The CYP3A4/PXR response element/xenobiotic-responsive enhancer module reporter vector was a gift from Dr. Bryan Goodwin (GlaxoSmithKline, Research Triangle Park, NC) and the pSG5-hPXR expression vector was provided by Dr. Steven Kliewer (University of Texas Southwestern Medical Center, Dallas, TX). Plasmids used in the mammalian two-hybrid assays were obtained from Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC). The pcDNA3-FLAG-hPXR wild-type (WT), W299D, and W299A expression vectors were obtained from Dr. Taosheng Chen (St. Jude Children’s Research Hospital, Memphis, TN). The cytomegalovirus/hemagglutinin/mPXR expression vector and tk-Cyp3a23-Luc reporter construct were obtained from Dr. Wen Xie (University of Pittsburgh, Pittsburgh, PA). Firefly luciferase activities were normalized with the pRL-TK Renilla luciferase plasmid from Promega.
Human Primary Hepatocyte and HepaRG Cell Cultures and Treatments.
Human hepatocytes were obtained from BioIVT (Baltimore, MD). Hepatocytes with ≥90% viability were seeded at 0.75 × 106 cells/well in 12-well biocoat plates in INVITROGRO CP Medium (BioIVT). After attachment at 37°C in a humidified atmosphere of 5% CO2, hepatocytes were cultured in complete Williams’ E medium and overlaid with Matrigel (0.25 mg/ml) as described previously (Li et al., 2008). Thirty-six hours after seeding, hepatocytes were treated with solvent (0.1% DMSO), CITCO (1 µM), RIF (10 µM), and PB (0.5 and 1 mM) or cotreated with SPA70 (2.5 µM) for 24 or 72 hours before harvesting cells for analysis of RNA or protein, respectively. Wild-type or CAR-KO HepaRG cells were seeded in 12-well plates at 1 × 105 cells/well and cultured for 21 days to induce differentiation according to Sigma-Aldrich’s instruction before initiation of the experiments.
Polymerase Chain Reaction Analysis.
Genomic DNA was isolated from WT- and hCAR-KO HepaRG cells using the QIAamp DNA Blood Mini Kit (QIAGEN, Germantown, MD) following the manufacturer’s instruction. With the deletion of TGGCCAGTAGG from exon 2 in the hCAR-KO HepaRG cells, specific primers for exon 2 (forward: 5′-AACACGTGACGTCATGGCCAG-3′; reverse: 5′-CCTCTGTTATGCCACCAGTT-3′) and exon 1 (forward: 5′-AAGCAGCAGCTTCCAATGAG-3′; reverse: 5-ACTCCTGGGCTCAAGCGATC-3′) were used for polymerase chain reaction (PCR) genotyping as described previously (Li et al., 2015).
RT-PCR Analysis.
Total RNA was isolated from cells using TRIzol reagent (ThermoFisher, Rockford, IL) and reverse transcribed to cDNA using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. RT-PCR assay was performed on an ABI StepOnePlus Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Mastermix (Qiagen). Primers for the human CYP2B6, CYP3A4, PXR, and glyceraldehyde-3-phosphate dehydrogenase include: CYP2B6, 5′-AGACGCCTTCAATCCTGACC-3′ and 5′-CCTTCACCAAGACAAATCCGC-3′; CYP3A4, 5′-GTGGGGCTTTTATGATGGTCA-3′ and 5′-GCCTCAGATTTCTCACCAACACA-3′; PXR, 5′-AAGCCCAGTGTCAACGCAG-3′ and 5′-GGGTCTTCCGGGTGATCTC-3′; and glyceraldehyde-3-phosphate dehydrogenase, 5′-CCCATCACCATCTTCCAGGAG-3′ and 5′-GTTGTCATGGATGACCTTGGC-3′. Induction values were calculated according to the following equation: fold over control = 2∆∆Ct, where ∆Ct represents the differences in cycle threshold numbers between the target gene and glyceraldehyde-3-phosphate dehydrogenase, and ∆∆Ct represents the relative change in these differences between control and treatment groups.
Western Blot Analysis.
Cell homogenate proteins (20 µg) were resolved on SDS-polyacrylamide gels (4%–12%) and electrophoretically transferred onto polyvinylidine fluoride membranes. Subsequently, membranes were blocked with 5% milk and incubated with antibodies against CYP2B6 (diluted 1:500; Abcam Inc., Cambridge, MA), CYP3A4 (1:5000; MilliporeSigma, Burlington, MA), hCAR (diluted 1:1000; Perseus Proteomics), or β-actin (1:5000; Sigma-Aldrich) at 4°C overnight. Blots were washed and incubated with horseradish peroxidase secondary antibodies, and developed with West Pico chemiluminescent substrates (ThermoFisher).
PXR Knockdown in Human Primary Hepatocytes.
Twenty-four hours after seeding, human primary hepatocytes were infected with negative control or small hairpin RNA against hPXR lentivirus particles that were packaged in human embryonic kidney 293T cells using the MISSION Lentiviral Packaging Mix System (Sigma-Aldrich). Infected hepatocytes were cultured in complete Williams’ E medium for 48 hours before treatment with solvent (0.1% DMSO), RIF (10 µM), and PB (1 mM) for 24 hours. Total RNA was prepared for RT-PCR analysis as described previously (Mackowiak et al., 2017).
Transient Transfection in HepG2 Cells.
HepG2 cells cultured in 24-well plates were transfected with different human or mouse PXR expression vectors with CYP3A4 or Cyp3a23 reporter constructs using X-tremeGENE 9 DNA Transfection Reagent (Roche Diagnostics Corporation, Indianapolis, IN). Twenty-four hours after transfection, cells were treated with 0.1% DMSO, RIF (10 µM), PCN (25 µM), and PB (0.1, 0.5 and 1 mM) for another 24 hours. In mammalian two-hybrid assays, HepG2 cells were transfected with reporter gene plasmid pG5luc, expression plasmids encoding GAL4-DBD/SRC-1 fusions, and VP16-AD/human or mouse PXR fusions for 24 hours before treatment with compounds as described previously. Subsequently, cell lysates were assayed for luciferase activities normalized against the activities of cotransfected Renilla luciferase using the Dual-Luciferase Kit (Promega). Data are represented as mean ± S.D. of three individual transfections.
Surface Plasmon Resonance Binding Assay.
Recombinant hPXR protein (Abcam Inc.) was covalently linked to the surface of a BIAcore CM5 sensor chip by direct immobilization with the amine coupling kit from GE Healthcare (Piscataway, NJ) following the manufacturer’s instructions. Purified glutathione S-transferase–fusion hCAR protein was captured by the monoclonal antibody against glutathione S-transferase that was bound to the surface of the BIAcore CM5 sensor chip as described previously (Zhang et al., 2010). RIF, PB and CITCO at indicated concentrations were used as analytes. The binding assay was carried out by injecting 60 μl each of the compounds in HEPES buffered saline surfactant P20 buffer (10 mM HEPES, pH 7.4, containing 150 mM NaCl and 0.05% P-20) with or without 2% DMSO for RIF, PB, and CITCO, respectively, at the flow rate of 30 μl/min at 25°C. The association and dissociation between analytes and hPXR or hCAR proteins were recorded respectively by SPR with a Biacore T200 (GE Healthcare) following the manufacturer’s instructions. Sensorgrams of the interaction generated by the instrument were analyzed using the software BIAeval 2.0 (GE Healthcare, Chicago, IL).
Molecular Modeling.
The crystal structure of the ligand-binding domains (LBDs) of hPXR [Protein Data Bank (PDB) ID: 1SKX] and hCAR (PDB ID: 1XVP) were retrieved from RCSB PDB (http://www.rcsb.org/)(Watkins et al., 2001; Xu et al., 2004). The chemical structures of PB, RIF, and CITCO were obtained from the ChemSpider structure database (http://www.chemspider.com). Prior to docking, the A-chain of 1SKX and D-chain of 1XVP were prepared in Discovery Studio 2019 (Dassault Systèmes BIOVIA, San Diego, CA) using the automatic prepare protein protocol, which removes ligands, adds missing hydrogens, and calculates side chain ionizations. The docking ligand structures were prepared using the automatic preparation tool to calculate the ionization state and canonical tautomer. The hPXR W299D and W299A mutation structures were generated via the build mutants protocol, which mutates the residues and then optimizes the conformation of neighboring residues. The binding sites were defined as the canonical ligand-binding cavities and the CDOCKER docking algorithm was used to find favorable ligand-protein poses by calculating the binding energy (ligand-protein interaction energy minus ligand strain). Reported binding energies were calculated using the calculate binding energies protocol as detailed previously (Wu et al., 2003).
Statistical Analysis.
All data represent the mean ± S.D. of at least three independent experiments. Statistical analyses included one-way related measures ANOVA followed by Tukey’s post-tests or two-way related measures ANOVA followed by Bonferroni post-tests where appropriate (GraphPad Prism 5.01). Statistical significance was set at *P < 0.05, **P < 0.01, or ***P < 0.001.
Results
PB Induces Both CYP2B6 and CYP3A4 with No Discernable Selectivity.
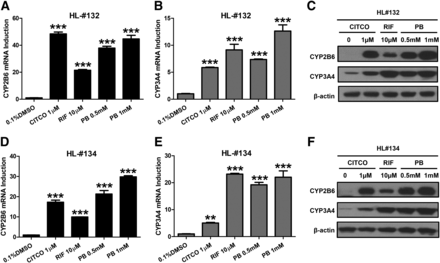
In human primary hepatocytes prepared from two liver donors (HL#132 and HL#134), we first investigated the effect of PB treatment on the mRNA and protein expression of CYP2B6 and CYP3A4, prototypical target genes for hCAR and hPXR, respectively. Selective activators of hCAR (CITCO) and hPXR (RIF) were used as positive controls. As expected, CITCO (1 µM) preferentially induces the expression of CYP2B6 over CYP3A4, while RIF (10 µM) exhibits pronounced induction of CYP3A4 over CYP2B6 (Fig. 1). At the concentrations of 0.5 and 1 mM, PB markedly induced both CYP2B6 and CYP3A4 without discernable preference. This induction pattern of PB clearly differs from that of either CITCO or RIF, supporting PB as a dual activator of both hCAR and hPXR.
PB induces CYP2B6 and CYP3A4 expressions in human primary hepatocytes. Human hepatocytes prepared from liver donors #132 (A–C) and #134 (D–F) were treated with 1 µM CITCO, 10 µM RIF, and 0.5 and 1 mM PB for 24 or 72 hours to analyze mRNA and protein, respectively. Expression of CYP2B6 and CYP3A4 mRNA and protein were measured using RT-PCR and western blotting assays. Results are expressed as mean ± S.D. (n = 3) (**P < 0.01; ***P < 0.001).
PB Induces CYP2B6 and CYP3A4 Expression in hCAR-KO HepaRG Cells.
HepaRG cells have been recognized as a useful alternative to human primary hepatocytes for in vitro metabolism and toxicology studies (Grime et al., 2010). A commercially available hCAR-KO HepaRG cell line obtained from Sigma-Aldrich provides a unique research tool for studying hCAR-independent gene transcription (Li et al., 2015). We first validated the hCAR-KO HepaRG cells using PCR genotyping and western blotting analysis. As expected, DNA from hCAR-KO HepaRG cells, lack of TGGCCAGTAGG in exon 2 (Fig. 2A), was not amplified by the specific exon 2 primers (Fig. 2B). This genetic modification led to the production of a nonfunctional hCAR protein that was barely picked up by the monoclonal hCAR antibody from Perseus Proteomics (Fig. 2C). Our subsequent results showed that PB robustly induced the expression of CYP2B6 and CYP3A4 in hCAR-KO HepaRG cells, while CITCO-mediated (a selective hCAR activator) induction of CYP2B6 was significantly attenuated at both mRNA and protein levels (Fig. 2, D and F). Interestingly, RIF, a selective hPXR activator, exhibits enhanced induction of CYP2B6 in hCAR-KO cells in comparison with WT HepaRG cells (Fig. 2, D, F, G, and I), suggesting loss of hCAR may relieve its competition with hPXR in provoking CYP2B6 expression. Collectively, these observations provide strong evidence that PB can induce CYP2B6 and CYP3A4 expression in a CAR-independent manner, most likely through the activation of hPXR.
PB induces P450 expression in HepaRG cells independent of CAR. (A) Schematic illustration of TGGCCAGTAGG deletion and primer localization for DNA genotyping of hCAR-KO HepaRG. After 21 days of differentiation, genomic DNA and cell homogenate prepared from both WT- and hCAR-KO HepaRG cells were subjected to PCR (B) and western blotting analysis (C). Differentiated WT- and hCAR-KO HepaRG cells were treated with 1 mM PB, 10 µM RIF, or 1 µM CITCO for 24 or 72 hours to measure CYP2B6 and CYP3A4 mRNA (D, E, G, and H) or protein expressions (F and I) using RT-PCR and western blotting assay, respectively. Results are expressed as mean ± S.D. (n = 3) (*P < 0.05; ***P < 0.001).
Inhibition of PXR Affects PB-Induced CYP3A4 Expression.
To further confirm the role of hPXR in PB-mediated induction, lentivirus small hairpin RNA was used to knock down hPXR expression in human primary hepatocytes. As shown in Fig. 3A, infection of lentiviral hPXR small hairpin RNA efficiently repressed the expression of endogenous hPXR in human primary hepatocytes at both mRNA and protein levels. The repression of hPXR subsequently results in marked attenuation of both PB- and RIF-medicated induction of CYP3A4 in human primary hepatocytes (Fig. 3B). In separate experiments, primary hepatocytes and hCAR-KO HepaRG cells were treated with PB or RIF in the presence or absence of SPA70, a newly identified selective hPXR deactivator (Lin et al., 2017). Notably, PB-induced expression of CYP3A4 in both hepatocytes and hCAR-KO HepaRG was significantly suppressed by SPA70 (Fig. 3, C and D). Together, these results strongly support that hPXR plays a pivotal role in PB-mediated CYP3A4 induction.
Inhibition of PXR affects PB-induced CYP3A4 expression. Human primary hepatocytes (HL#139) were infected with lentiviral/hPXR/small hairpin RNA (shRNA) or lentiviral-negative control followed by treatment with 0.1% DMSO, 10 µM RIF, or 1 mM PB as detailed in Materials and Methods. Expression of hPXR (A) and CYP3A4 (B) was analyzed by RT-PCR and western blotting assays. In a separate experiment, human primary hepatocytes (HL#139) and fully differentiated hCAR-null HepaRG cells were treated with 0.1% DMSO, 10 µM RIF, 1 mM PB, and 2.5 µM SPA70 alone or cotreatment of SPA70 with RIF or PB (C and D). RT-PCR and western blotting were used to measure the mRNA and protein expression of CYP3A4. Data are expressed as mean ± S.D. (n = 3) (***P < 0.001).
PB Activates hPXR but Not mPXR.
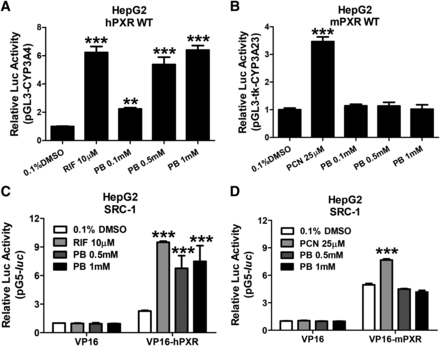
PXR is a ligand-driven transcription factor that stimulates gene transcription by recognizing, binding, and recruiting coactivators to the responsive element–containing promoters of target genes. Here, we first investigated the ability of PB to activate hPXR and mPXR in cell-based luciferase reporter assays. HepG2 cells were transfected with CYP3A luciferase reporter constructs in the presence of hPXR or mPXR expression vectors, respectively. As demonstrated in Fig. 4A, hPXR was robustly activated by PB (0.1, 0.5, and 1 mM) in a concentration-dependent manner. In contrast, PB at the same concentrations did not active mPXR (Fig. 4B). As expected, RIF and PCN as selective hPXR and mPXR agonists markedly activated their respective target receptor. Given that agonistic binding of PXR often leads to enhanced recruitment of coactivator SRC-1 to the transcription complex (Takeshita et al., 2001; Gollamudi et al., 2008), we subsequently tested whether PB could recruit SRC-1 to PXR by mammalian two-hybrid assays. After treatment with RIF 10 µM or PB at 0.5 and 1 mM, the interaction between hPXR and SRC-1 was significantly enhanced (Fig. 4C). On the other hand, PB does not change the interaction between mPXR and SRC-1, while PCN (25 µM) increased the binding of SRC-1 to mPXR as expected (Fig. 4D). These observations suggest that PB selectively activates hPXR but not mPXR by increasing the interaction of hPXR and coactivator SRC-1.
PB selectively activates hPXR but not mPXR by increasing the interaction of hPXR with coactivator SRC-1. HepG2 cells were transfected with hPXR (A) or mPXR (B) in the presence of CYP3A4/PXR response element/xenobiotic-responsive enhancer module or tk-Cyp3a23-Luc reporter constructs. Transfected cells were then treated with 0.1, 0.5, and 1 mM PB for 24 hours. RIF (10 µM) and PCN (25 µM) were used as positive controls for hPXR and mPXR, respectively. Luciferase activities were determined and expressed relative to vehicle control (0.1% DMSO). Mammalian two-hybrid assays were performed in HepG2 cells transiently transfected with the reporter gene plasmid pG5luc and expression plasmids encoding GAL4-DBD/SRC-1 in the presence of VP16-hourPXR (C) or VP16-mPXR (D) fusion proteins. Cells were treated with 0.1% DMSO, 0.5 and 1 mM PB, 10 µM RIF, or 25 µM PCN for an additional 24 hours before determination of luciferase activities. Three independent measures from each treatment were analyzed and are expressed as mean ± S.D. (**P < 0.01; ***P < 0.001).
PB Binds Directly to hPXR.
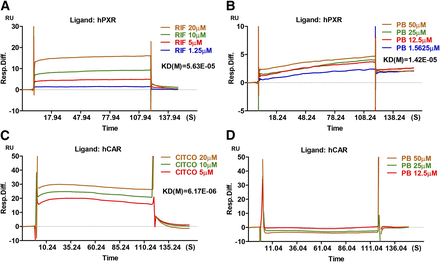
PB is known to activate CAR through a well-characterized indirect mechanism involving dephosphorylation of CAR without direct ligand binding (Negishi, 2017). To explore the potential interaction between PB and hPXR, we next characterized the kinetics of PB binding to hPXR using a SPR binding assay. As expected, the known agonists of hPXR (RIF) and hCAR (CITCO) bind to their respective target proteins robustly with KD values of 5.63 × 10−05 and 6.17 × 10−06, respectively (Fig. 5, A and C). We found that PB exhibits efficient binding to hPXR (KD = 1.42 × 10−05) but not to hCAR (Fig. 5, B and D). The lack of PB binding to hCAR in this assay not only attested to PB-mediated indirect activation of CAR, it also validated the specificity of the PB-hPXR interaction.
Surface plasmon resonance sensograms of hPXR interaction with PB. BIACORE SPR affinity assays were carried out to measure the comparative binding kinetics of RIF (A, positive control) and PB (B) to hPXR protein as described in Materials and Methods. Measurements of comparative binding kinetics of CITCO (C) and PB (D) to hCAR protein were used as positive and negative controls, respectively. Sensorgrams of the interaction generated by the instrument were analyzed by the software BIAeval 3.2.
Computational Modeling of PB with hPXR.
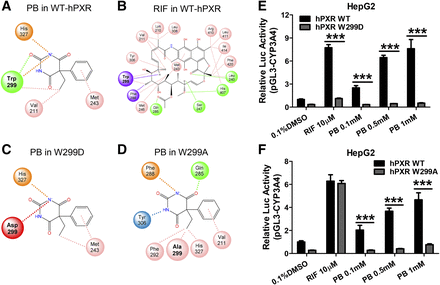
To understand the structural basis of PB binding to hPXR, docking analysis was carried out based on the crystal structure of the hPXR LBD (PDB ID: 1SKX) using the CDOCKER algorithm within Discovery Studio 2019. Docking of PB into the LBD of hPXR reveals that PB interacts with a relatively small number of amino acids in the binding pocket, while with a noticeably strong association with W299 via multiple favorable interactions (Fig. 6A). On the other hand, RIF with a significantly larger molecular weight is associated with many amino acid residues including W299 in the large and flexible LBD of hPXR (Fig. 6B). Further analysis of the total binding energy indicated that both PB and RIF bind efficiently with hPXR with free energies of −6.8 and −20.1 kcal/mol, respectively. Given the predicted strong interaction of PB with W299, we next analyzed the potential binding interaction changes when tryptophan at 299 was mutated to either the negatively charged aspartic acid (W299D) or a neutral residue alanine (W299A). These mutations have resulted in markedly reduced association with PB in the binding cavity (Fig. 6, C and D) and increased free energies of +5.7 and +0.5 kcal/mol, respectively. Additional luciferase reporter assays using WT-, W299D-, and W299A-hPXR constructs confirmed the critical role of W299 in PB-mediated activation of hPXR, where W299D mutant drastically disrupted both RIF and PB activation of hPXR (Fig. 6E), W299A, on the other hand, selectively eliminated PB- but not RIF-mediated hPXR activation (Fig. 6F). Furthermore, to corroborate the docking method used, CITCO and PB were docked in the hCAR LBD (PDB ID: 1XVP), in which they are known direct and indirect hCAR activators, respectively. The results support CITCO as a favorable hCAR agonist (−17.0 kcal/mol), while PB has minimal interaction with hCAR (−0.1 kcal/mol) (Supplemental Fig. 1). Detailed three-dimensional diagrams illustrating docking of PB and RIF in the LBD of WT- W299D-, and W299A-hPXR are presented as Supplemental Material (Supplemental Fig. 2). Together, these findings suggest that PB is an agonist of hPXR, and W299 in the ligand-binding pocket of hPXR plays a critical role in PB’s binding and activation of hPXR.
Docking analysis of PB interaction with WT and mutants of W299 hourPXR. Binding poses of PB and RIF with the WT-hPXR (A and B) as well as PB with W299D and W299A mutation of hPXR (C and D) are illustrated in a two-dimensional conformation. Favorable interactions with amino acids in the PXR binding pocket, indicated by dashed lines, were determined using CDOCKER. Green circles with dashed lines are hydrogen bonds, pink circles with dashes are alkyl interactions, purple circles with dashes are π-σ interactions, blue circles are π-hydrogen interactions, orange circles with dashes are π-anion interactions, and red circles with dashes are unfavorable interactions. In a separate experiment, HepG2 cells were transfected with the WT-, W299D- (E) or W299A-hPXR (F) in the presence of CYP3A4/PXR response element/xenobiotic-responsive enhancer module-Luc reporter constructs. Cells were then treated with 0.1, 0.5, and 1 mM PB, 10 µM RIF, or 0.1% DMSO for 24 hours. Luciferase activities were determined and expressed relative to vehicle control. Three independent measures from each treatment were analyzed and are expressed as mean ± S.D. (***P < 0.001).
Discussion
PB has been used extensively as a model compound studying CAR-dependent induction of cytochrome P450 (P450) genes across different species. In particular, the indirect (ligand-independent) nature of PB-mediated activation of CAR has led to improved understanding of CAR as a signaling molecule (Mutoh et al., 2013; Yang and Wang, 2014). It is well known that CAR and PXR, two closely related xenobiotic sensors, can regulate each other’s target genes through crosstalk; many drugs are able to modulate the activity of both nuclear receptors, making interpretation of pharmacological responses associated with CAR/PXR complicated. The fact that PB-induced expression of Cyp2b10 and Cyp3a11 is completely abolished in CAR-null mice unequivocally established PB as an activator of CAR but not PXR in mice (Wei et al., 2000). However, our knowledge is limited regarding the role of PB in hPXR regulation. Using a hCAR-KO HepaRG cell line, our study shows that PB robustly induces the expression of both CYP2B6 and CYP3A4 through a hCAR-independent pathway, and further inhibition of hPXR fully eliminated this induction. Mechanistic investigation demonstrated that PB selectively activates hPXR by enhancing the functional interaction between SRC-1 and hPXR but not mPXR. Specifically, we show that PB directly binds to hPXR in key association with W299 in the ligand-binding pocket and mutation of this amino acid functionally disrupts PB-mediated hPXR activation. Our results provide novel mechanistic insights into PB as a dual activator of both hPXR and hCAR (Fig. 7).
Schematic illustration of PB as a dual activator of hPXR and hCAR. PB activates hPXR and hCAR through direct ligand binding and indirect mechanisms, respectively.
CYP2B6 and CYP3A4 are prototypical transcriptional targets for hCAR and hPXR, respectively. However, accumulating evidence reveals that crosstalk between CAR and PXR results in reciprocal transactivation of CYP2B6 and CYP3A genes (Xie et al., 2000). The asymmetric crossregulation of these genes has led to observations where selective activation of hCAR by CITCO preferentially induces the expression of CYP2B6 over CYP3A4, while activation of hPXR by RIF leads to induction that favors CYP3A4 over CYP2B6 (Faucette et al., 2007). In the current study, PB markedly induced the expression of both CYP2B6 and CYP3A4 in human hepatocytes with less discernible differences, a pattern that is noticeably unlike that of CITCO or RIF. It is noteworthy that this phenomenon also differs from previous studies in which pronounced induction of Cyp2B over Cyp3A was observed in PB-treated rats and mice (Jones and Lubet, 1992; Ariyoshi et al., 2001), supporting the notion that PB, a rodent CAR activator, functions as a dual activator of both CAR and PXR in human.
Recently, well-differentiated HepaRG cells have been established as a valuable surrogate for human primary hepatocytes, in which these cells express important liver-enriched transcriptional factors including CAR and PXR and exhibit efficient drug induction of major P450 enzymes and transporters (Grime et al., 2010; Jackson et al., 2016). Using hCAR-KO HepaRG cells, we found that PB markedly induces both CYP2B6 and CYP3A4, while CITCO-mediated induction of CYP2B6 was nearly abolished when compared with WT-HepaRG cells, suggesting that PB coordinates hCAR-independent transactivation of these P450 genes. It is worth mentioning that although CITCO is widely accepted as a selective hCAR agonist and exhibits preferential induction of CYP2B6 over CYP3A4, it also activates hPXR with relatively low efficacy (Maglich et al., 2003; Auerbach et al., 2005; Faucette et al., 2006). This may explain why hCAR-KO drastically reduced CITCO-mediated induction of CYP2B6, but only had negligible effects on CITCO-mediated rather weak induction of CYP3A4. Most recently, SPA70 has been reported as a potent and selective hPXR antagonist with low cytotoxicity (Lin et al., 2017). Using this compound, we showed that PB-induced expression of CYP3A4 was completely eliminated in hCAR-KO HepaRG cells when cotreated with SPA70. Collectively, these findings indicate hPXR is the transcriptional factor that is responsible for the PB-mediated P450 induction in hCAR-KO HepaRG cells.
Although PB is known as a universal CAR activator and induces P450 genes among different species, it is now evident that PB regulates human and mouse PXR differently. The luciferase reporter assays conducted in this study revealed that PB concentration dependently activates hPXR but not mPXR. These observations are in agreement with our own as well as a number of other previous reports indicating PB is able to transactivate hPXR in cell-based reporter assays (Wang et al., 2004; Pinne et al., 2017). Mechanistically, agonistic binding and activation of PXR involve the release of preoccupied corepressors, such as silencing mediator of retinoid and thyroid receptors, and the recruitment of coactivators such as SRC-1 to the PXR/retinoid X receptor heterodimer (Watkins et al., 2003; Gollamudi et al., 2008). In contrast, indirect activation of nuclear receptors such as CAR by PB often translocates the constitutively activated CAR to the nucleus without further enhancing its already existing interaction with coactivators (Kanno et al., 2014; Yang et al., 2014). Our results from a mammalian two-hybrid assay demonstrate that PB efficiently recruits SRC-1 to the DNA-protein complex of hPXR while not to mPXR, implying that PB may activate hPXR through direct agonistic binding.
To gain insight into the mechanistic basis of PB-mediated activation of hPXR, we subsequently evaluated the binding capacity of PB to hPXR in SPR assays. Notably, PB exhibits effective hPXR binding even at concentrations (12.5–50 µM) far below the concentrations typically used for P450 induction (0.25–1 mM) in human primary hepatocytes (Faucette et al., 2004; Rotroff et al., 2010). These findings were supported by the expected lack of binding of PB to hCAR at similar concentrations. The potent binding of RIF and CITCO to hPXR and hCAR, respectively, further validates the effectiveness of this experiment. These findings also led us to explore the structure-activity nature of this PB-hPXR interaction. The reported X-ray crystal structure of the hPXR LBD has allowed us to generate reliable computational models by docking different agonists (Watkins et al., 2001). As the largest hPXR agonist identified thus far, RIF interacts with numerous amino acids within the LBD in our docking studies, in a pattern that is similar to a previous report (Chrencik et al., 2005). Importantly, while PB interacts only with a small number of amino acids within the ligand-binding pocket of hPXR, it exhibits a robust interaction with W299, a key component of a cluster of hydrophobic residues inside the ligand-binding cavity. A number of studies have reported that W299 is a conserved amino acid that involves hydrophobic interaction with various ligands (Watkins et al., 2001, 2003; Chrencik et al., 2005). Recently, Banerjee et al. (2016) showed that replacing W299 with differently charged residues significantly reduced agonistic activation of hPXR. Moreover, the same mutation may alter hPXR activity differentially in a ligand-specific manner. Based on these findings, we next probed the interaction between PB and hPXR bearing a negatively charged aspartic acid W299D or a neutral residue alanine W299A mutation, both known to alter ligand-mediated hPXR activation. Docking of PB to hPXR-W299D drastically reduced its interaction with the W299 site by increasing the free energy from a favorable −6.8 kcal/mol in WT-hPXR to an unfavorable +5.7 kcal/mol in the W299D mutant. Our luciferase reporter experiments showed that with the negatively charged residue, the W299D mutant exhibited loss of function to both PB and RIF treatment. Conversely, the hPXR-W299A mutant, with a PB binding free energy of +0.5 kcal/mol, displayed ligand-specific inhibition of PB- but not RIF-mediated hPXR activation. This finding concurs with Banerjee’s report, where W299A selectively reduces hPXR activation by T0901317 but not by SR12813 or RIF, which are all robust activators of WT-hPXR (Banerjee et al., 2016).
In conclusion, we provide convincing evidence to show that PB, a selective mouse CAR activator, is a dual activator of both hCAR and hPXR. Our results demonstrate that PB activates hPXR through direct ligand binding, enhancing its recruitment of coactivator SRC-1. Further evidence reveals that PB specifically interacts with the W299 residue in the ligand-binding pocket of hPXR, and mutation of W299 to the negatively charged amino acid D299 or the neutral residue A299 fully disrupted PB-mediated hPXR activation. While the current study focuses specifically on PB-mediated P450 induction, given the expanded roles of PXR in energy metabolism and cell growth (Gupta et al., 2008) it is tempting to speculate that differential regulation of hPXR and mPXR may also contribute to other species-specific biologic discrepancies stimulated by PB treatment, including its roles in energy homeostasis and cancer development.
Acknowledgments
We acknowledge provision of hPXR-W299D and hPXR-W299A plasmids by Dr. Taosheng Chen (Saint Jude Children’s Research Hospital, Memphis, TN), the CYP3A4-PXR response element/xenobiotic-responsive enhancer module reporter vector by Dr. Bryan Goodwin (GlaxoSmithKline, Research Triangle Park, NC), the pSG5-hPXR expression vector by Dr. Steven Kliewer (University of Texas Southwestern Medical Center, Dallas, TX), and the cytomegalovirus/hemagglutinin/mPXR expression and tk-Cyp3a23-Luc reporter constructs by Dr. Wen Xie (University of Pittsburgh, Pittsburgh, PA). We are also grateful to Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC) for providing plasmids used in the mammalian two-hybrid assays, Dr. Yinghua Zhang (University of Maryland School of Medicine, Baltimore, MD) for assistance in the plasmon resonance binding analysis, and Dassault Systèmes BIOVIA for providing a software license for BIOVIA Discovery Studio 2019.
Authorship Contributions
Participated in research design: L. Li, Welch, Z. Li, Mackowiak, Wang.
Conducted experiments: L. Li, Welch, Z. Li, Mackowiak, Heyward.
Contributed new reagents or analytic tools: Heyward.
Performed data analysis: L. Li, Welch, Mackowiak, Swaan, Wang.
Wrote or contributed to the writing of the manuscript: L. Li, Welch, Z. Li, Mackowiak, Heyward, Swaan, Wang.
Footnotes
- Received March 5, 2019.
- Accepted July 7, 2019.
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
This work was supported by the National Institutes of Health National Institute of General Medicine [Grants GM107058 and GM121550] and National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK61425]. Bryan Mackowiak was partly supported by The University of Maryland’s Center of Excellence in Regulatory Science and Innovation Scholars Program funded by the Food and Drug Administration [Project 1U01FD005946].
↵
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
Abbreviations
- CAR
- constitutive androstane receptor
- CITCO
- 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- hCAR
- human constitutive androstane receptor
- hPXR
- human pregnane X receptor
- KO
- knockout
- LBD
- ligand-binding domain
- mPXR
- mouse pregnane X receptor
- P450
- cytochrome P450
- PB
- phenobarbital
- PCN
- pregnenolone 16α-carbonitrile
- PCR
- polymerase chain reaction
- PDB
- Protein Data Bank
- PXR
- pregnane X receptor
- RIF
- rifampicin
- RT-PCR
- real-time polymerase chain reaction
- SPA70
- 4-((4-(tert-butyl)phenyl)sulfonyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole
- SPR
- surface plasmon resonance
- SRC-1
- steroid receptor coactivator-1
- WT
- wild type
- Copyright © 2019 by The American Society for Pharmacology and Experimental Therapeutics